Abstract
Ion channels gated by cyclic nucleotides have crucial roles in neuronal excitability and signal transduction of sensory neurons. Here, we studied ligand binding of a cyclic nucleotide-activated K+ channel from Mesorhizobium loti and its isolated cyclic nucleotide-binding domain. The channel and the binding domain alone bind cyclic AMP with similar affinity in a non-cooperative manner. The cAMP sensitivities of binding and activation coincide. Thus, each subunit in the tetrameric channel acts independently of the others. The binding and gating properties of the bacterial channel are distinctively different from those of eukaryotic cyclic nucleotide-gated channels.
Introduction
Ion channels activated by cyclic nucleotides fall into two subfamilies: cyclic nucleotide-gated (CNG) channels, and hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels, (for reviews, see Kaupp & Seifert, 2001, 2002; Robinson & Siegelbaum, 2003; Craven & Zagotta, 2006). Both types of channel carry a carboxy-terminal cyclic nucleotide-binding domain (CNBD). However, two notable differences distinguish CNG and HCN channels from each other. CNG channels require cyclic nucleotides to open, whereas HCN channels activate on hyperpolarization and their activity is modulated by cyclic nucleotides. Furthermore, HCN channels are cAMP-selective and are sensitive to cAMP concentrations in the nanomolar range, whereas CNG channels are cGMP-selective and activate at micromolar concentrations of cGMP. The molecular interactions that tune affinity and selectivity of cyclic nucleotide binding are not fully understood because ligand affinity has not been measured directly for any CNG or HCN channel. Instead, binding affinities of CNG channels have been inferred from electrophysiological studies (Gordon & Zagotta, 1995a, 1995b; Varnum et al, 1995). As binding and gating are intimately coupled, it is difficult to separate one from the other (Colquhoun, 1998).
Here, we studied ligand binding of a bacterial cyclic nucleotide-activated K+ channel from Mesorhizobium loti (mlCNG; Clayton et al, 2004; Nimigean et al, 2004) and its isolated CNBD by using isothermal titration calorimetry and fluorescence techniques.
Results
Preparation of a cAMP-free CNBD
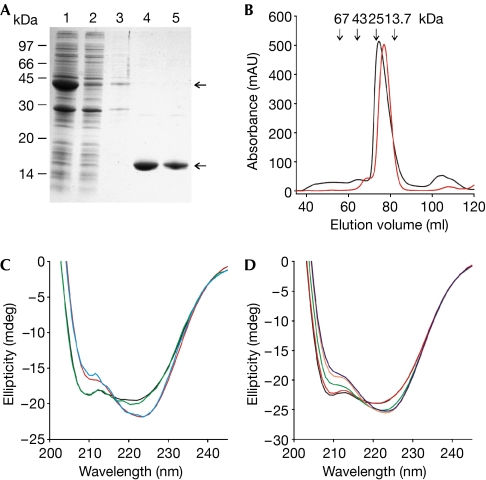
The CNBD was expressed and purified as a glutathione-S-transferase (GST)-fusion protein (Fig 1A). In size-exclusion chromatography, the CNBD was eluted in a single peak (at approximately 20 kDa) close to the expected molecular mass (Mw) of the monomer (15 kDa; Fig 1B). The purified CNBD contains tightly bound cAMP that cannot be removed by size-exclusion chromatography or extensive dialysis (Clayton et al, 2004). To remove cAMP, we denatured and refolded the CNBD protein. The cAMP content was analysed by reversed-phase high-performance liquid chromatography (RP-HPLC; see supplementary Fig 1 online). The mean cAMP content was 84.2±12.4% (n=25) before denaturation and below the detection limit after refolding.
Figure 1.
Purification and characterization of the CNBD protein. (A) The CNBD–GST fusion protein was expressed in Escherichia coli and affinity purified (lane 1: column input; lane 2: non-bound; lane 3: wash; lanes 4 and 5: elution of the CNBD protein after thrombin cleavage). Upper and lower arrows indicate the CNBD–GST fusion protein and the cleaved CNBD protein, respectively. Coomassie blue staining of an SDS–polyacrylamide gel. (B) Gel-filtration profile of the purified CNBD protein. Red trace: elution profile of the native CNBD protein; black trace: profile of the CNBD protein after denaturation and refolding. Arrows indicate the elution volume of marker proteins. (C) CD spectra of the purified CNBD protein (8.2 μM) before unfolding (blue), after refolding in the absence of cAMP (black), after refolding in the presence of cAMP (15 μM, red) and of the R348A mutant in the absence of cAMP (green). (D) CD spectra of the refolded CNBD protein (11.4 μM) titrated with cAMP at different concentrations (black, 0 μM; red, 5 μM; green, 10 μM; orange, 15 μM; blue, 20 μM). CD, circular dichroism; CNBD, cyclic nucleotide-binding domain; GST, glutathione-S-transferase.
The circular dichroism (CD) spectra of the cAMP-bound form and the cAMP-free form were significantly different (Fig 1C), suggesting that a conformational change occurs on cAMP binding. A similar change in CD spectra was reported for protein kinase A and G (Johnson & Wong, 1989; Landgraf et al, 1990). Fig 1D shows CD spectra of the refolded protein at various cAMP concentrations. The spectra reflect the transition from the cAMP-free to the cAMP-bound conformation. At saturating cAMP concentrations, the CD spectrum of the refolded protein becomes almost identical to that of the protein before unfolding (Fig 1C). We also compared the CD spectra of the refolded CNBD with a mutant (R348A) that has a lower cAMP affinity and can therefore be prepared in a cAMP-free form without denaturation/refolding (Clayton et al, 2004). The CD spectra of the R348A mutant and the refolded, cAMP-free CNBD were almost identical (Fig 1C), suggesting that both CNBDs adopt the same conformation. We analysed the CD spectra to estimate the changes in secondary structure that occur on ligand binding (supplementary Fig 2, and supplementary Tables 1 and 2 online). The helical content adopts slightly higher values in the ligand-bound form compared with the ligand-free form. The differences were most pronounced when we compared the helical content of the refolded CNBD in the presence of saturating cAMP (45.5%) with that of the R348A mutant (40%). The overall helical content derived from the crystal structures of the wild-type CNBD and the R348A CNBD is lower (36% and 30 %, respectively; supplementary Table 3 online) than predicted from the CD spectra; however, the difference in the helical content of the two proteins is in a similar range.
Non-cooperative cyclic nucleotide binding of CNBD
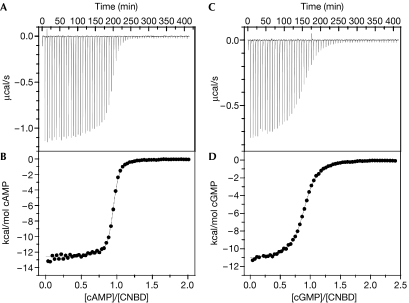
The binding of cyclic nucleotides to the CNBD was studied by isothermal titration calorimetry (ITC; Wiseman et al, 1989). After injection of cAMP into a sample cell, the CNBD–ligand interaction was recorded as a change in heat (Fig 2A), from which the dose–response relationship was derived (Fig 2B). A similar dose–response relationship was obtained for cGMP (Fig 2C,D). Analysis of the data using a single binding-site model yielded a mean KD of 107±11 nM for cAMP (n=4) and of 499±69 nM (n=3) for cGMP. We also determined the KD values for several 8-substituted analogues of cyclic nucleotides (Table 1). These analogues activate mammalian CNG channels at significantly lower concentrations than either cAMP or cGMP (Kaupp & Seifert, 2002). All cyclic nucleotide analogues bind to the CNBD; notably, 8-CPT-cAMP binds with higher affinity than cAMP, whereas 8-CPT-cGMP has a lower affinity than cGMP (Table 1). Thus, the binding cavity of CNBD is distinctively different from the site in mammalian CNG channels. The stoichiometry of binding to the CNBD for all cyclic nucleotides tested was 0.95±0.11 (n=22), showing that all CNBD molecules participate in binding and that the fraction of misfolded CNBD is negligible.
Figure 2.
Isothermal titration calorimetry of cyclic-nucleotide binding to the CNBD protein. (A) Heat changes during successive injections of 4 μl cAMP (600 μM) to the CNBD protein (56.7 μM) in a sample cell of 1.4 ml. (B) Binding curve. The peaks in (A) have been integrated, normalized to the cAMP concentration, and plotted against the molar ratio of cAMP to CNBD protein. The solid line represents a nonlinear least-squares fit to a single-site binding model. The KD value was 107 nM; the binding stoichiometry was 0.94. (C,D) Similar experiments as in (A,B), but with cGMP as the ligand. Successive 4 μl injections of cGMP (465 μM) to the CNBD protein (36.7 μM). The KD value was 495 nM; the binding stoichiometry was 0.89. CNBD, cyclic nucleotide-binding domain.
Table 1.
Summary of binding constants
| CNBD | R348A | mlCNG | |||||
|---|---|---|---|---|---|---|---|
| Ligand | ITC | Stoichiometry | 8-NBD-cAMP fluorescence | Trp fluorescence | 8-NBD-cAMP fluorescence | 8-NBD-cAMP fluorescence | Rb+-flux assay |
| KD (nM) | KD (nM) | KD (nM) | KD (μM) | KD (nM) | K1/2 (nM) | ||
| cAMP | 107±11 (4) | 0.98±0.12 | 67.8±8.7 (7) | 80 (4) | 18.5±4.3 (7) | 81.6±17.5 (6) | 60–100*,† |
| 8-Br-cAMP | 127±11 (3) | 0.95±0.06 | — | — | — | — | |
| 8-CPT-cAMP | 34±25 (3) | 0.91±0.08 | — | — | — | — | |
| 8-NBD-cAMP | 24±1.5 (3) | 0.84±0.10 | 22.0±10.9 (11) | — | 7.3 (1) | 15.9±2.7 (5) | |
| cGMP | 499±69 (3) | 0.97±0.08 | 300.4±15.4 (7) | — | 22.3±5.7 (6) | 320.7±25.5 (6) | 600* |
| 8-Br-cGMP | 236±23 (3) | 1.1±0.06 | — | — | — | — | |
| 8-CPT-cGMP | 2,770±70 (3) | 0.88±0.1 | — | — | — | — | |
CNBD, cyclic nucleotide-binding domain; mlCNG, cyclic nucleotide-gated channel from Mesorhizobium loti; ITC, isothermal titration calorimetry; Trp, tryptophan; *Nimigean et al, 2004; †Clayton et al, 2004
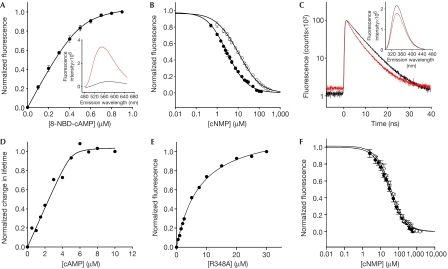
We used the fluorescent analogue 8-NBD-cAMP as an independent measure of ligand binding (Fig 3A). The fluorescence of the NBD group is enhanced on binding to a protein (Fig 3A, inset; Kraemer et al, 2001). The dose-dependent increase of 8-NBD-cAMP fluorescence was well described using a simple binding model (see equations (1–3), supplementary information online). The KD value was 22.0±10.9 nM (n=11), in agreement with the KD value obtained using ITC (Table 1). The KD values for binding of cAMP and cGMP were derived from competition experiments using equations (4–7), (supplementary information online; Fig 3B). The KD values were 67.8±8.7 nM (n=7) for cAMP and 300.4±15.4 nM (n=7) for cGMP, again in agreement with the ITC measurements (Table 1).
Figure 3.
Ligand binding to the CNBD protein by fluorescence spectroscopy. (A) Increase of 8-NBD-cAMP fluorescence (emission at 550 nm) on binding to the CNBD protein (0.5 μM). 8-NBD-cAMP fluorescence in the absence of the CNBD protein was subtracted. The solid line represents a nonlinear least-squares fit to ΔF=RL · x (equation (3), see supplementary information online). The KD value was 17.6 nM. Inset: emission spectrum of 8-NBD-cAMP (1 μM) in the absence (black) and presence (red) of CNBD protein (1 μM). (B) Competition between cAMP (closed circles) or cGMP (open circles) and 8-NBD-cAMP (1 μM) for binding to the CNBD (1 μM). Solid lines represent a nonlinear least-squares fit to ΔF=RLf · x (equation (7), see supplementary information online). The KD values were 73.5 nM (cAMP) and 296.9 nM (cGMP). (C) Decay of fluorescence of the tryptophan (Trp) residue of CNBD (5 μM) in the absence (black) and presence (red) of cAMP. Absolute values of lifetime were as follows: 0 μM cAMP (A1=12,252; τ1=6.778 ns; A2=3,133; τ2=2.035 ns; τ̄=6.357 ns); 10 μM cAMP (A1=10,999; τ1=6.258 ns; A2=3,889; τ2=2.185 ns; τ̄=5.674 ns). Inset: emission spectrum of Trp in the absence (black) and presence (red) of cAMP. CNBD concentration was 5 μM. (D) Normalized changes in fluorescence lifetime of Trp as a function of cAMP concentration. The solid line represents a nonlinear least-squares fit to equation (3) (supplementary information online). The KD value was 80 nM. (E) Increase of fluorescence of 8-NBD-cAMP (0.5 μM) on binding to increasing concentrations of the mutant CNBD (R348A). The KD value was 7.3 μM. (F) Competition between cAMP or cGMP and 8-NBD-cAMP for binding to mutant CNBD (R348A, 3 μM). The KD values were 22.9 μM (cAMP) and 27.7 μM (cGMP). CNBD, cyclic nucleotide-binding domain.
The CNBD carries a single tryptophan (Trp) residue in the αA′ helix of the short C-linker region preceding the binding domain. When cAMP was added to the ligand-free CNBD, the lifetime of Trp fluorescence was considerably shortened (Fig 3C) and the intensity of Trp fluorescence was markedly reduced (Fig 3C, inset). cAMP did not shift the emission spectrum, arguing that fluorescence is quenched by a nearby phenylalanine (Lackowicz, 1999; sequence in one letter code: 223FVRNW227). The αA′ helix becomes bent in the cAMP-bound form (Clayton et al, 2004), which is consistent with the idea that the distance between Trp and Phe shortens, resulting in enhanced quenching. We determined the lifetime of Trp fluorescence at various concentrations of cAMP (Fig 3D). Fitting of equation (3) (see supplementary information online) to the dose–response relationship yielded a KD value of 80 nM for cAMP, which is in agreement with the KD values obtained by ITC and fluorescence spectroscopy.
R348 is responsible for high-affinity ligand binding
HCN channels have an arginine (Arg) residue in the αC-helix of the CNBD that interacts with cAMP and a nearby glutamic acid (Glu) residue (Zagotta et al, 2003). The Arg residue is also present in the M. loti channel (R348), but is lacking in CNG channels. We compared the binding of 8-NBD-cAMP, cAMP and cGMP to wild type and the R348A mutant (Fig 3E,F). The binding affinity of the mutant was significantly lower with KD values in the micromolar range (8-NBD-cAMP: 7.3 μM; cAMP: 18.5±4.3 μM (n=7); cGMP: 22.3±5.7 μM (n=6)). This finding indicates that the high cAMP sensitivity is conferred by the interaction of this Arg residue with the ligand. If this interaction is lacking, the affinity becomes similar to the K1/2 of activation of CNG channels.
Ligand binding to CNBD and the entire channel is similar
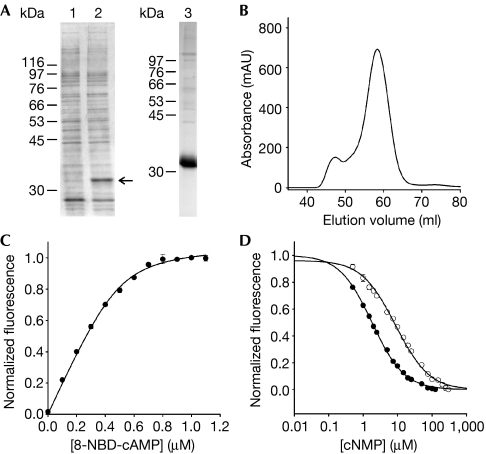
In the crystal structure, the CNBD forms a dimer with significant areas of contact, and the dimer interface has been proposed to be involved in channel gating (Clayton et al, 2004). To examine the importance of this interaction for ligand binding, we determined ligand affinity for the tetrameric full-length mlCNG. The mlCNG protein was expressed in Escherichia coli with a C-terminal hexahistidine tag (Nimigean et al, 2004). In SDS–polyacrylamide gel electrophoresis, the protein showed an apparent Mw of 32 kDa (Fig 4A). The mlCNG protein was purified through a Co2+-affinity column (Fig 4A). The homogeneity of the purified protein was tested by size-exclusion chromatography. Most of the mlCNG protein eluted in a single peak (Fig 4B). A small fraction eluted in the void volume and probably represents aggregated material. Similarly to the CNBD, the purified mlCNG protein contained cAMP (53.3±14.1%; n=15). As the denaturation/refolding procedure is less suitable for membrane proteins, we used 8-CPT-cGMP to displace cAMP from the channel protein. Owing to its lower affinity (Table 1), 8-CPT-cGMP can be removed easily by washing. Thereafter, the mlCNG protein contained virtually no cAMP or 8-CPT-cGMP (see supplementary Fig 1 online).
Figure 4.
Ligand binding of the full-length mlCNG protein. (A) mlCNG detected by Coomassie blue staining (arrow) in bacterial lysates (lane 1: no induction; lane 2: IPTG induction). mlCNG protein purified by Co2+-affinity chromatography (lane 3). (B) Size-exclusion chromatography of the purified mlCNG protein. Most of the mlCNG protein eluted at 58.3 ml. (C) Normalized increase of fluorescence of 8-NBD-cAMP on binding to mlCNG protein (5 μM). 8-NBD-cAMP fluorescence in the absence of the mlCNG protein was subtracted. The solid line represents a non-linear least-squares fit to Δ F=RL · x (equation (3), see supplementary information online). The KD value was 17.3 nM. (D) Competition between 8-NBD-cAMP (0.5 μM) and cAMP (closed circles) or cGMP (open circles) for binding to the mlCNG protein (0.3 μM). The solid lines represent a nonlinear least-squares fit to Δ F=RL · x (equation (7), see supplementary information online). The KD values were 71.3 nM (cAMP) and 363.9 nM (cGMP). IPTG, isopropyl-β-D-thiogalactopyranoside; mlCNG, Mesorhizobium loti cyclic nucleotide-gated channel; 8-NBD-cAMP, 8-[[2-[(7-Nitro-4-benzofurazanyl)amino]ethyl]thio]adenosine–3′,5′-cyclic monophosphate.
At the high concentrations required for ITC, the cAMP-free mlCNG protein is prone to form aggregates. Therefore, we studied the binding of cyclic nucleotides by 8-NBD-cAMP fluorescence. Fig 4C shows the increase of fluorescence on binding of 8-NBD-cAMP to mlCNG. The mean KD value was 15.9±2.7 nM (n=5), which is similar to the value obtained for the isolated CNBD (Table 1). The KD values for binding of cAMP and cGMP were determined by a competition assay (Fig 4D). The KD value was 81.6±17.5 nM (n=6) for cAMP and 320.7±25.5 nM (n=6) for cGMP. In conclusion, the KD values for binding of cyclic nucleotides to mlCNG and the CNBD are very similar.
Discussion
We studied ligand binding in a cyclic nucleotide-activated K+ channel and its isolated CNBD. The mlCNG channel binds cAMP with high affinity that is similar to the K1/2 value of channel activation (Clayton et al, 2004; Nimigean et al, 2004). Furthermore, the cyclic-nucleotide selectivity (cAMP>cGMP) for both binding and activation is similar (Table 1; Nimigean et al, 2004). Both binding and activation are non-cooperative, indicating that the binding event is not affected by intersubunit contact and that binding sites in the tetrameric channel act independently of each other. This conclusion is supported by the finding that binding affinities of the monomeric CNBD and the tetrameric full-length mlCNG channel are virtually identical. This finding is surprising for the following reason. In the crystal, the CNBD forms a dimer with extensive areas of contact in the αA′ helix of the linker region (Clayton et al, 2004). On ligand binding, the two αA′ helices slide in opposite directions along the interface, suggesting that the dimer interface has a crucial role in the gating process. We reasoned that some of the binding energy would be used for this gating transition and that the binding affinity of the channel should therefore be lower than that of the isolated CNBD. However, our results indicate that the main conformational change for the closed–open transition occurs in the CNBD and the linker themselves, and that the gating step involving the rearrangement of adjacent linker regions and the subsequent movement of S6 are energetically less demanding.
In this respect, activation of bacterial and mammalian CNG channels seems to be completely different. Mammalian CNG channels activate cooperatively, a process that requires allosteric coupling between binding and gating, and involves inter- and intrasubunit contact sites. In fact, many mutations outside the CNBD—for example, in the amino terminus, the pore region and the C-linker—profoundly affect ligand sensitivity and selectivity (for reviews, see Kaupp & Seifert, 2002; Craven & Zagotta, 2006). Complex allosteric models, including the Monod–Wyman–Changeux model (Goulding et al, 1994) and the coupled dimer model (Liu et al, 1998), have been used to account for the cooperative behaviour (for a review, see Li et al, 1997). These models assume that all binding sites are equivalent and that the ligand affinity is higher in the open state than in the closed state by an allosteric factor f (Li et al, 1997; however, see Li & Lester, 1999). The differences among CNG channels in ligand sensitivity and selectivity have been largely attributed to gating rather than binding (Li et al, 1997). While this manuscript was being revised, Biskup et al (2007) showed that binding sites are not equivalent, and that both positive and negative cooperativity exist between A2 subunits of the olfactory CNG channel (also see Nache et al, 2005). In particular, these authors conclude that the binding of the second ligand almost entirely promotes the closed–open transition (Biskup et al, 2007), whereas the third and fourth ligands drive the channel away from the second binding event, thereby stabilizing the open state. Our observation, that activation of the mlCNG channel closely follows ligand binding, raises the intriguing possibility that the mlCNG channel might be fully opened on binding of a single cAMP molecule. Thus, the difference between mammalian and bacterial CNG channels might be in the cooperativity of binding. The mlCNG channel is resistant to functional expression in eukaryotic cells (W.B. & R.S., unpublished data), which has impeded its characterization. If this obstacle can be overcome, the above hypothesis can be rigorously tested experimentally.
For the same reason, we have been unable to confirm whether mlCNG is a genuine CNG channel. In fact, two features suggest that mlCNG is more similar to HCN than CNG channels. First, modulation of HCN channel activity by cAMP is essentially non-cooperative (Gauss et al, 1998). Second, the cAMP affinity of mlCNG is of the same order as that of the HCN2 channel (F. Winkhaus & R.S., unpublished data), but it is approximately 100-fold higher than the cyclic-nucleotide affinity of CNG channels inferred from electrophysiological studies (Varnum et al, 1995; Gordon & Zagotta, 1995a, 1995b). Residue R348 in the αC helix of the mlCNG-CNBD is conserved in HCN, but not in CNG channels. Exchange of R348 by a neutral residue lowers the cAMP affinity by 200- to 400-fold, but does not abolish cAMP binding completely. In fact, the binding affinities of the R348A mutant for cyclic nucleotides are similar to the K1/2 value of activation of CNG channels (Kaupp & Seifert, 2002). Thus, R348 is a crucial residue that sets apart high- and low-affinity binding cavities by providing a site of contact with cAMP in addition to those sites shared by HCN and CNG channels, including the phosphate-binding cassette (Zagotta et al, 2003; Clayton et al, 2004).
Methods
Materials. Cyclic nucleotides were obtained from Biolog (Bremen, Germany) and Sigma-Aldrich (München, Germany). N-decyl-β-D-maltopyranoside (DM) was obtained from Calbiochem (Schwalbach, Germany).
Protein expression and purification. The DNA encoding a mlCNG was cloned from genomic DNA. The gene was modified at the 3′-end to encode a hexahistidine (His)6 tag. For expression in E. coli (BL21 (DE3) pLysE), the gene was subcloned into the pET-11a vector (Novagen, Schwalbach, Germany). Expression was induced at A600∼0.4 with 0.6 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 4 h at 20°C. Cells were lysed by sonification in lysis buffer I (in mM): 295 NaCl, 50 Na+ phosphate, 5 KCl, 2 MgCl2, pH 8.0 containing DNaseI (25 μg/ml) and a protease inhibitor cocktail (Complete, Roche, Mannheim, Germany). After centrifugation (93,000g, 20 min at 4°C), the pellet was solubilized for 2 h at 4°C in lysis buffer II (in mM): 295 NaCl, 20 Na+ phosphate, 5 KCl, 2 MgCl2, 5 imidazole, 20 DM, 20% (v/v) glycerol, pH 8.0 and DNaseI (25 μg/ml). The solubilized protein was purified using Co2+-affinity chromatography (HiTrap and HisTrap, GE Healthcare, Freiburg, Germany). The protein was eluted with a buffer containing (in mM) 295 NaCl, 20 Na+ phosphate, 5 KCl, 5 DM, 20% (v/v) glycerol and 5-500 imidazole, pH 8.0. Gel filtration was carried out using a Superdex-200 column (Prepgrade 16/60 Hiload, GE Healthcare) in a buffer containing (in mM) 100 KCl, 20 Na+ phosphate and 5 DM, pH 8.0. The protein concentration was determined with Bradford (Bio-Rad, München, Germany).
The CNBD was expressed and purified as a GST-fusion protein (Clayton et al, 2004). The coding region for residues 216–355 of the mlCNG protein was cloned into pGEX-2T (GE Healthcare). For expression, the DNA was transformed into BL21-(DE3)pLysE cells. Overnight expression (20°C) was induced at A600∼0.4 with 0.6 mM IPTG.
Cells were lysed in PBS buffer, and the soluble and insoluble fractions were separated by centrifugation. The supernatant was passed over a glutathione-Sepharose 4B column, followed by incubation with thrombin for cleavage of the CNBD domain from the GST tag overnight. Cleaved protein was eluted and further purified by size-exclusion chromatography on a Superdex 75 column (Prepgrade 16/60 Hiload, GE Healthcare).
Removal of cAMP from the CNBD and mlCNG protein. To remove residual cAMP from the CNBD, the protein was denatured on a PD10 column equilibrated with 6 M guanidine hydrochloride. The buffer was exchanged several times until cAMP was completely removed. The denatured cAMP-free CNBD was refolded by rapid dilution into a solution containing (in mM) 100 NaCl, 10 Na+ phosphate, 5 glutathione (reduced), 0.5 glutathione (oxidized), 0.5 L-arginine and 10 Na+ EDTA, pH 7.0.
To remove residual cAMP from mlCNG, the protein bound to the Co2+-affinity column was incubated three times with the low-affinity ligand 8-CPT-cGMP (1 mM), followed by washing steps with 20 ml washing buffer (in mM) 295 NaCl, 20 Na+ phosphate, 5 KCl, 10 imidazole and 5 DM, pH 8.0. Subsequently, the column was washed with 200 ml washing buffer; the protein was eluted with a solution containing (in mM) 295 NaCl, 20 Na+ phosphate, 5 KCl, 400 imidazole and 5 DM, pH 8.0. The cyclic-nucleotide content of protein was determined by HPLC, as described in the supplementary information online.
Binding experiments using fluorescence techniques. The binding of 8-NBD-cAMP was recorded with a fluorescence spectrophotometer (QM-4; PTI, Birmingham, NJ, USA) at 22±2°C. 8-NBD-cAMP was excited at 470 nm and the emission spectra were collected between 480 nm and 650 nm. The cAMP analogue and the mlCNG were suspended in standard buffer containing (in mM) 100 KCl, 20 Na+ phosphate and 2.5 DM, pH 8.0. Binding measurements with the CNBD were carried out in a solution without detergent containing (in mM) 100 KCl and 10 Na+ phosphate, pH 7.4. In competition experiments, binding of 8-NBD-cAMP to the mlCNG or CNBD was studied in the presence of different concentrations of cAMP or cGMP.
Fluorescence lifetime. Time-resolved decay of fluorescence was measured with a fluorescence spectrophotometer (Fluotime100, Picoquant, Berlin, Germany) using a pulsed LED (centre wavelength: 298 nm; pulse width: 550 ps) as an excitation source. Emission was observed at 340 nm. The fluorescence decay curve was reconstructed by repetitive measurements of the time between the excitation pulse and the emitted photon using time-correlated single-photon counting (Kaneko et al, 2002). The time resolution was less than 500 ps. Decay curves were analysed with Fluofit4 (Picoquant, Berlin, Germany) using:
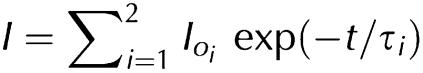 |
where Ioi are the respective intensities and τi the characteristic lifetimes. The average lifetime τ̄ was calculated as:
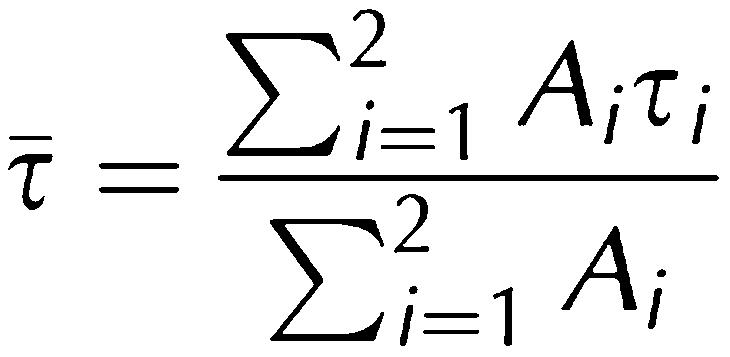 |
where Ai is the relative amplitude.
CD spectra. Far-UV spectra were recorded on a Jasco J-810 spectropolarimeter at 22±2°C (0.2 cm path-length cuvette) in 20 mM Na+ phosphate buffer at pH 7.4. The CD spectra were analysed for secondary structure elements as described in the supplementary information online.
Isothermal titration calorimetry. Measurements were carried out at 25°C using a VP-ITC microcalorimeter (MicroCal, Northampton, MA, USA; volume of sample cell 1.4 ml). The reference cell contained water. The cyclic nucleotides and the CNBD protein were suspended in PBS. The CNBD protein (36–85 μM) was titrated with cAMP (400–1,200 μM) and cGMP (250–987 μM) using injections of 4–5 μl. Calorimetric data were analysed with the Origin Software (MicroCal; version 5.0) using equations described previously (Wiseman et al, 1989).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v8/n8/extref/7401025-s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Bott for support with the ITC measurements, T. Kaneko (Kazusa DNA Research Institute) for M. loti DNA, J. Schmitz for technical help, and A. Baumann and S. Bungert for reading the manuscript.
References
- Biskup C, Kusch J, Schulz E, Nache V, Schwede F, Lehmann F, Hagen V, Benndorf K (2007) Relating ligand binding to activation gating in CNGA2 channels. Nature 446: 440–443 [DOI] [PubMed] [Google Scholar]
- Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH (2004) Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell 119: 615–627 [DOI] [PubMed] [Google Scholar]
- Colquhoun D (1998) Binding, gating, affinity and efficacy: the interpretation of structure–activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125: 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven KB, Zagotta WN (2006) CNG and HCN channels: two peas, one pod. Annu Rev Physiol 68: 375–401 [DOI] [PubMed] [Google Scholar]
- Gauss R, Seifert R, Kaupp UB (1998) Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393: 583–587 [DOI] [PubMed] [Google Scholar]
- Gordon SE, Zagotta WN (1995a) Localization of regions affecting an allosteric transition in cyclic nucleotide-activated channels. Neuron 14: 857–864 [DOI] [PubMed] [Google Scholar]
- Gordon SE, Zagotta WN (1995b) A histidine residue associated with the gate of the cyclic nucleotide-activated channels in rod photoreceptors. Neuron 14: 177–183 [DOI] [PubMed] [Google Scholar]
- Goulding EH, Tibbs GR, Siegelbaum SA (1994) Molecular mechanism of cyclic-nucleotide-gated channel activation. Nature 372: 369–374 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Wong SS (1989) Conformational changes of type II regulatory subunit of cAMP-dependent protein kinase on cAMP binding. Febs Lett 247: 480–482 [DOI] [PubMed] [Google Scholar]
- Kaneko H, Putzier I, Frings S, Gensch T (2002) Determination of intracellular chloride concentration in dorsal root ganglion neurons by fluorescence lifetime imaging. Curr Top Membr 53: 167–189 [Google Scholar]
- Kaupp UB, Seifert R (2001) Molecular diversity of pacemaker ion channels. Annu Rev Physiol 63: 235–257 [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Seifert R (2002) Cyclic nucleotide-gated ion channels. Physiol Rev 82: 769–824 [DOI] [PubMed] [Google Scholar]
- Kraemer A, Rehmann HR, Cool RH, Theiss C, De Rooji J, Bos JL, Wittinghofer A (2001) Dynamic interaction of cAMP with the Rap guanine-nucleotide-exchange factor Epac1. J Mol Biol 306: 1167–1177 [DOI] [PubMed] [Google Scholar]
- Lackowicz JR (1999) Principles Fluorescence Spectroscopy 2nd edn, pp 446–485. New York, NY, USA: Kluwer Academic/Plenum [Google Scholar]
- Landgraf W, Hofmann F, Pelton JT, Huggins JP (1990) Effects of cyclic GMP on the secondary structure of cyclic dependent protein kinase and analysis of the enzyme's amino-terminal domain by far-ultraviolet circular dichroism. Biochemistry 29: 9921–9928 [DOI] [PubMed] [Google Scholar]
- Li J, Lester HA (1999) Functional roles of aromatic residues in the ligand-binding domain of cyclic nucleotide-gated channels. Mol Pharmacol 55: 873–882 [PubMed] [Google Scholar]
- Li J, Zagotta WN, Lester HA (1997) Cyclic nucleotide-gated channels: structural basis of ligand efficacy and allosteric modulation. Q Rev Biophys 30: 177–193 [DOI] [PubMed] [Google Scholar]
- Liu DT, Tibbs GR, Paoletti P, Siegelbaum SA (1998) Constraining ligand-binding site stoichiometry suggests that a cyclic nucleotide-gated channel is composed of two functional dimers. Neuron 21: 235–248 [DOI] [PubMed] [Google Scholar]
- Nache V, Schulz E, Zimmer T, Kusch J, Biskup C, Koopmann R, Hagen V, Benndorf K (2005) Activation of olfactory-type cyclic nucleotide-gated channels is highly cooperative. J Physiol 569: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean CM, Shane T, Miller C (2004) A cyclic nucleotide modulated prokaryotic K+ channel. J Gen Physiol 124: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RB, Siegelbaum SA (2003) Hyperpolarization-activated cation currents: from molecules to physiological function. Annu Rev Physiol 65: 453–480 [DOI] [PubMed] [Google Scholar]
- Varnum MD, Black KD, Zagotta WN (1995) Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron 15: 619–625 [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin L-N (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179: 131–137 [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425: 200–205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information