Abstract
The complete mitochondrial DNA (mtDNA) (16,896 nt) of the wallaroo (Macropus robustus) was sequenced. The concatenated amino acid sequences of 12 mitochondrial protein-coding genes of the wallaroo plus those of a number of other mammals were included in a phylogenetic study of early mammalian divergences. The analysis joined monotremes and marsupials (the Marsupionta hypothesis) to the exclusion of eutherians. The analysis rejected significantly the commonly acknowledged Theria hypothesis, according to which Marsupialia and Eutheria are grouped together to the exclusion of Monotremata. The region harboring the gene for lysine tRNA (tRNA-Lys) in the mtDNA of other vertebrates is in the wallaroo occupied by a sequence (tRNA-Lys) that lacks both an anticodon loop as well as the anticodon for the amino acid lysine. An alternative tRNA-Lys gene was not identified in any other region of the mtDNA of the wallaroo, suggesting that a tRNA-Lys of nuclear origin is imported into marsupial mitochondria. Previously described RNA editing of tRNA-Asp and rearrangement of some tRNA genes were reconfirmed in the mtDNA of the wallaroo. The divergence between Monotremata/Marsupialia and Eutheria was timed to ≈130 million years before present (MYBP). The same calculations suggested that Monotremata and Marsupialia diverged ≈115 MYBP and that Australian and American marsupials separated ≈75 MYBP. The findings also show that many, probably most, extant eutherian orders had their origin in middle to late Cretaceous times, 115–65 MYBP.
Keywords: mammalian evolution, Marsupionta hypothesis, dating of evolutionary divergences, RNA editing, RNA import
It is commonly acknowledged that the evolutionary relationship among the three main mammalian groups, monotremes, marsupials, and eutherians, has been conclusively resolved by traditional approaches. An early divergence of the monotremes is generally postulated on the basis of synapomorphic characters shared by marsupials and eutherians (1–4). This traditional view is known as the Theria hypothesis. Based on morphological findings, only three papers have questioned this relationship, instead advocating a sister-group relationship between marsupials and monotremes, the Marsupionta hypothesis (5–7). While the characters used by Gregory (5) have been rejected as being plesiomorphic and thus not suitable for phylogenetic reconstruction, the characters of identical tooth replacement in marsupials and monotremes studied by Kühne (6, 7) have been considered to be convergent (1, 8, 9).
The relationships among monotremes, marsupials, and eutherians have been studied on the basis of molecular data of both nuclear and mitochondrial genes (10–13). Neither the analysis of myoglobin (10) or mitochondrial 12S rRNA (12), both of which were based on a rather limited amount of data, did resolve the relationship, whereas a study of the P1 gene, based on 70 aa (11), supported the Theria hypothesis. Analyses of the protein-coding genes of complete mitochondrial molecules, mtDNAs, of 10 vertebrate species, including a monotreme, the platypus, and an American marsupial, the opossum, have provided strong support for the Marsupionta hypothesis (13).
In the present study we examine the phylogenetic relationship among Monotremata, Marsupialia, and Eutheria by including an Australian marsupial, the wallaroo, Macropus robustus. This inclusion breaks up the long marsupial branch, thereby reducing the potential effect of long branch attraction. In addition, availability of the mtDNA of the wallaroo permitted dating of the evolutionary divergence between Australian and American marsupials.
Studies of the mtDNA of the opossum, Didelphis virginiana, have shown that the tRNA genes around the origin of light strand replication have been rearranged and an atypical secondary structure of tRNA-Lys was also reported (14). The presently described complete mtDNA of the wallaroo has made it possible to examine both these features, in particular whether a functional tRNA-Lys has been transpositioned to a different location in the mitochondrial genome. The mtDNA of the wallaroo also made it possible to investigate if the RNA editing of the anticodon for tRNA-Asp, where a cytosine is edited to uridine (15, 16), is characteristic for both Australian and American marsupials.
MATERIALS AND METHODS
Enriched mtDNA was isolated from frozen liver of the wallaroo applying a previously described approach (17). The tissue sample was donated by Bengt Röken (Kolmården Zoo, Kolmården, Sweden). Restriction fragments generated by single or combined digestions with different restriction enzymes (BamHI, BclI, BglI, HindIII, BlnI, NheI, SpeI, XbaI) were ligated into M13 and cloned in Escherichia coli JM101 (18). The cloned fragments covered the entire molecule. Sequencing was according to the dideoxy termination technique (19) with [α-35S]dATP, using both universal and numerous specific oligonucleotide primers.
The mtDNA sequence of the wallaroo has been deposited in the EMBL database (accession number Y10524Y10524). Users of the sequence are kindly requested to refer to the present paper and not only to the accession number of the sequence. For the phylogenetic analysis the protein coding genes from the following species were aligned to the wallaroo sequence: carp, X61010X61010 (20); loach, M91245M91245 (21); frog, X02890X02890 (22); chicken, X52392X52392 (23); platypus, X83427X83427 (13); opossum, Z29573Z29573 (14); hedgehog, X88898X88898 (24); mouse, J01420J01420 (25); rat, X14848X14848 (26); guinea pig (27); rabbit (C. Gissi, personal communication); cat, U20753U20753 (28); gray seal, X72004X72004 (29); harbor seal, X63726X63726 (30); horse, X79547X79547 (31); donkey, X97337X97337 (32); Indian rhinoceros, X97336X97336 (33); cow, V00654V00654 (34); fin whale, X61145X61145 (17); blue whale, X72204X72204 (35); gibbon (36); Sumatran orangutan, X97707X97707 (37); Bornean orangutan, D38115D38115 (38); gorilla, X93347X93347 (39); pygmy chimpanzee, D38116D38116 (38); human (African), D38113D38113 (38); and human (Caucasian, “Lund”), X93334X93334 (40).
Gaps and ambiguous alignments adjacent to gaps were excluded from the phylogenetic analyses. The NADH6 gene, which is encoded by the opposite strand relative to the remaining protein-coding genes, and which differs significantly in nucleotide and amino acid composition, was not included in the data set. The phylogenetic analyses were performed using the phylip (41), molphy (42), and puzzle (43) packages. For the neighbor-joining (NJ) (44) and maximum parsimony (MP) (45) analyses, confidence values for internal branches were established by bootstrapping. This was not necessary for maximum likelihood (ML) (46) analysis, as quartet puzzling (QP) provides the corresponding reliability values.
RESULTS AND DISCUSSION
General Features of the Genome.
The mtDNA of the wallaroo is 16,896 nt long and encodes the 13 protein-coding genes and the two (12S and 16S) rRNAs characterizing metazoan mtDNAs. The molecule, however, has only 21 typical tRNAs as compared with 22 in all other vertebrate mtDNAs studied so far. The start codon of NADH5 is ATT rather than ATG or ATA (methionine). This is consistent with the notion that ATT/ATC start codons may specify methionine (47). Four genes, COII, COII, NADH3, and NADH4, have incomplete termination codons (T). Stop codons of this kind are completed to functional stop codons by posttranscriptional polyadenylylation (48).
The control region is 1428 nt long. Unlike many other mammals, it does not contain repetitive motifs. Three conserved sequence blocks (CSBs), which are believed to be involved in the replication of the molecule, have been described in the mitochondrial genome of mammals (49). In the wallaroo two of these, CSBII (location, 16,592–16,609) and CSBIII (location, 16,644–16,662) are similar to other mammalian CSBs, whereas CSBI is probably missing or much less conserved than CSBII and CSBIII. A termination-associated sequence, presumably involved in the termination of replication in mammalian mtDNAs (50), was identified in position 15,736–15,774. The pronounced sequence conservation of both CSBs and termination-associated sequences in virtually all mammals (51) supports their hypothesized function.
In the presently described Australian marsupial the two tRNAs for serine show unusual features (Fig. 1). The tRNA-Ser(AGY) lacks the DHU arm, a feature conserved among all vertebrates, and the anticodon stem of tRNA-Ser(UCN) consists of 6 bp instead of 5 as found in other mitochondrial tRNAs. Also in the tRNA-Ser(UCN) only 1 nt is found between the acceptor stem and the DHU stem, a feature that has been described for eutherians and for the opossum, but which is lacking in the monotreme and other vertebrates (13, 52).
Figure 1.
Inferred secondary structure of atypical tRNAs in the mtDNA of the wallaroo.
The tRNA for Aspartic Acid.
The primary and the inferred secondary structures of the mitochondrial tRNA-Asp gene of the wallaroo conform with those of other mammals, while the canonical anticodon sequence GTC is lacking (Fig. 1). Thus a typical tRNA-Asp is missing in the mtDNA of the wallaroo. Instead of the typical anticodon GTC, the tRNA of the wallaroo has the anticodon GCC, which in metazoans would recognize glycine codons. It has been shown that unedited tRNA-Asp may function as a complementary tRNA-Gly in marsupial mitochondria (53), and that in the mitochondria of marsupials tRNA-Asp(GCC) is edited at the second anticodon position (from C to U) to restore its original function (14–16). Because a probable RNA editing of tRNA-Asp has been described in a New Guinean marsupial (15), it is conceivable that in marsupials in general the tRNA-Asp is edited to restore its function as an aspartate carrying tRNA.
The tRNA for Lysine.
The tRNA-Lys(UUU) gene is located between the protein-coding genes COII and ATPase8 in tetrapod mtDNAs. In this position in the opossum a sequence resembling a tRNA-Lys with some unusual features in primary and secondary structures has been reported (14). Because atypical tRNAs are not uncommon in mitochondria (54, 55), these features were not detailed when the mitochondrial genome of the opossum was described (14).
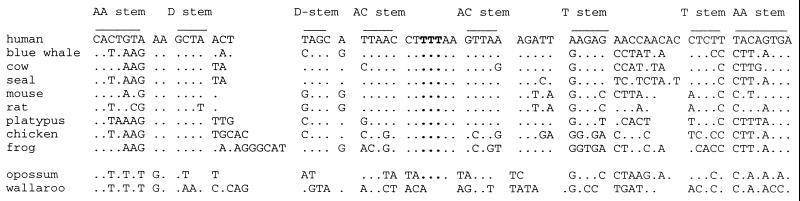
The length of vertebrate tRNA-Lys is 64–74 nt. Despite the length differences, the characteristics of tRNA-Lys have been conserved with respect to both the primary sequence and inferred secondary structure. In the wallaroo the sequence occupying this region (“tRNA-Lys”) is 64 nt long. As evident from Fig. 2, there are striking differences in the primary structure between the two marsupials and other vertebrate sequences. These differences extend even into regions that are generally conserved in vertebrates. The inferred secondary structure of the wallaroo, “tRNA-Lys” (shown in Fig. 1) is not tRNA-like, notably because the absence of an anticodon loop that in typical tRNA-Lys genes contains 7 nt, including an anticodon T triplet. Furthermore, all metazoan mitochondrial tRNA genes (except tRNA-Met) are characterized by a T occurring at the 5′ position adjacent to the anticodon. This feature is missing in both marsupials. Despite the pronounced difference between typical mammalian tRNA-Lys genes and the corresponding marsupial region, it is conceivable that the “tRNA-Lys” originates from a functional tRNA-Lys gene, although the proper tRNA function has been lost. It may thus be a pseudogene. The 3′ and 5′ ends of “tRNA-Lys” are to a considerable extent conserved between the two marsupials. This region shows compensatory substitutions and the sequence can be transformed into a tRNA-like structure. It has been postulated that tRNAs and hairpin-loop structures in mtDNA may serve as recognition sites for the processing of the polycistronic transcript (48). It is thus probable that a function of this kind has been maintained in the “tRNA-Lys.”
Figure 2.
Alignment of the mitochondrial “tRNA-Lys” sequences from different mammals and the tRNA-Lys of the wallaroo. The different stem regions are overlined, the anticodon sequence is in boldface type. In the wallaroo sequence no anticodon (TTT) or anticodon loop can be identified.
A potential tRNA-Lys gene was not identified at any other location in the mtDNA of either the opossum or the wallaroo. The absence of a functional tRNA-Lys would make mitochondrial translation impossible. In case RNA-editing was responsible for reforming a “tRNA-Lys” transcript to a functional tRNA-Lys, a large amount of RNA editing would be required. Although it has been shown that extensive editing of mitochondrial tRNAs occurs in protzoan mitochondria (56) and that nucleotides are added to the 3′ end of a mitochondrial tRNA in monotremes (57), these editing events are confined to the acceptor stems.
To generate a typical mitochondrial tRNA-Lys from the wallaroo “tRNA-Lys” gene by means of RNA editing, a minimum of five base changes in different regions of the tRNA would be required, in addition to four uridine insertions into the anticodon loop. Sequencing of cDNAs of marsupial mitochondrial lysosyl-tRNAs does not suggest editing (M. Dörner, personal communication) and therefore, although RNA import into mammalian mitochondria is still subject to debate (58), we find it likely, on the basis of the absence of a canonical tRNA-Lys gene in the mtDNA of the wallaroo, that the translational function in marsupial mitochondria is accomplished by import of a nuclear encoded tRNA-Lys rather than by RNA editing.
PHYLOGENY
The various methods used in phylogenetic reconstruction have different advantages (and drawbacks) in establishing the correct tree, and in some instances the results may differ depending on the method used. To minimize the methodological risk we have applied the three most commonly used methods of tree reconstruction, namely MP (45), NJ (44), and ML (46). We also applied a new algorithm called QP (43), which, on the basis of a ML estimation, examines all possible quartets of taxa in the process of tree reconstruction.
The analysis was based on the concatenated sequences, both nucleotide and amino acid, of 12 protein-coding genes (excluding NADH6). The nucleotide sequences were analyzed with respect to all changes at the 1st codon position except leucine codon transitions (all synonymous), all changes at the 2nd codon position, and all transversions at the 3rd codon position.
The reliability of phylogenetic reconstruction—i.e., the potential to find the true tree—may differ depending on how well the set of characters conforms with the assumptions made by the algorithms used (42, 59, 60). Base composition can vary among taxa (60), and this may affect the outcome of the analysis. Differences in nucleotide composition will also influence the amino acid composition, if the compositional bias extends to nonsynonymous changes.
The initial vertebrate alignment included 29 vertebrate sequences: two fishes, one frog, one bird, the platypus, two marsupials, and 22 eutherians. The length of the alignment after excluding gaps and ambiguous sites was 3248 aa. The mean values for the amino acid frequencies conform with the expected values used by the ML algorithm of the molphy (42) and puzzle (43) program packages.
With respect to amino acid frequencies the most conspicuous deviations from the mean values were observed in the fishes, the chicken, and the hedgehog. According to a χ2-test, the sequences of these species deviate significantly (at the 95% level) from the mean amino acid frequency and consequently from that of the ML expectation. To avoid an effect by compositional bias on the analysis the deviating species were excluded from the phylogenetic reconstruction leaving only the frog as an outgroup.
At the nucleotide level, the second codon position of the fishes, the chicken, and the hedgehog fail the χ2-test for homogeneity of base composition, whereas at the 1st codon position 11 of the 29 sequences fail to show such homogeneity. However, when synonymous changes at the 1st codon position were excluded, only the two fishes, the chicken, and the hedgehog deviated significantly from the mean values. Compared with other species the chicken has preference for C and G over A and T. This is particularly pronounced at the 3rd codon position at which transversions for 8 aa are synonymous. This bias occurs also at the 2nd and 1st codon positions, leading to the deviations observed at the amino acid level. The hedgehog sequence, however, is biased toward a higher frequency of A or T rather than C or G, a situation that is opposite to that in the chicken.
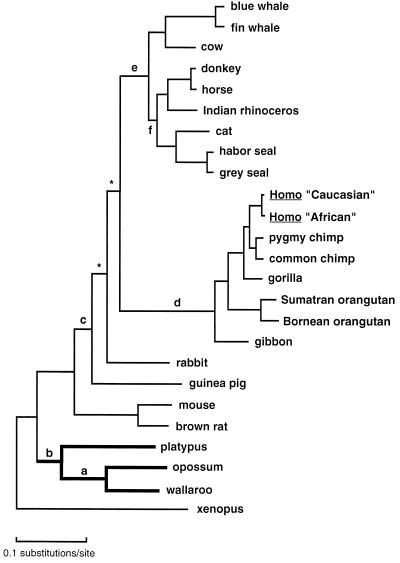
All phylogenetic reconstruction methods of both amino acid and nucleotide sequences resulted in the topology shown in Fig. 3, which shows a ML tree based on amino acid sequences and the mtREV-22 model of amino acid evolution. The QP algorithm left no quartet unresolved among the 12,650 that were analyzed to establish this tree. Branch “b” which joins the monotreme and the marsupials (Marsupionta hypothesis) received highly significant (99.9%) support. Other data sets and different reconstruction methods favored this relationship as well (Table 1). The bootstrap values and QP support for selected branches of particular phylogenetic interest are shown in Table 1. All phylogenetic reconstruction methods of all types of data sets, except the MP analysis of all codon positions combined, yielded strongest support for the Marsupionta tree. In many cases this support was statistically significant at or above the 95% confidence level. Some analyses of reduced data sets provided less support, underlining the importance of using comprehensive data in phylogenetic analyses.
Figure 3.
Maximum likelihood tree (42) of concatenated amino acid sequences of 12 H-strand encoded mitochondrial genes of 25 vertebrates. The mtREV-22 model (42) was used for the ML computations. The topology represents the Marsupionta tree, with marsupials and monotremes as sister groups. Branch lengths are according to genetic distance. Bootstrap and QP support values for branches a–f are shown in Table 1.
Table 1.
Support values of selected branches
| a | b | c | d | e | f | |
|---|---|---|---|---|---|---|
| NJ | ||||||
| 1 | 100 | 68.2 | 64.8 | 100 | 100 | 64.4 |
| 2 | 100 | 91.6 | (85.4) | 100 | 100 | 47.3 |
| 12 | 100 | 95.2 | 94.1 | 100 | 100 | 52.2 |
| 123 | 100 | 79.2 | 94.2 | 100 | 100 | 81.2 |
| aa | 100 | 100 | 74 | 100 | 100 | 94 |
| MP | ||||||
| 1 | 100 | 39.3 | 80.0 | 100 | 100 | — |
| 2 | 100 | 69.1 | (77.5) | 100 | 99.3 | — |
| 12 | 100 | 46.6 | 97.3 | 100 | 100 | 48.5 |
| 123 | 100 | 31.0 | 93.7 | 100 | 100 | — |
| aa | 100 | 77.5 | 80.5 | 100 | 100 | 56.2 |
| ML/QP | ||||||
| 1 | 96.0 | 94.3 | (69.4) | 100 | 95.1 | 74.7 |
| 2 | 98.7 | 96.9 | (83.6) | 99.0 | 99.1 | — |
| 12 | 98.4 | 94.8 | (76.1) | 100 | 98.3 | 70.3 |
| 123 | 97.6 | 95.8 | (77.9) | 100 | 99.0 | 67.7 |
| aa | 99.9 | 99.9 | (64.6) | 100 | 91.2 | 89.1 |
The numbers show bootstrap and support values in percent for selected branches (a–f) shown in Fig. 3. The analyses of the assembly of 25 taxa were carried out by NJ, MP, and ML/QP (maximum likelihood using the quartet puzzling) tree search algorithm for 1st (1) 2nd (2), 1st plus 2nd (12), 1st plus 2nd plus 3rd codon positions (123), as well as amino acid (aa), sequences. For 1st codon position all changes except transitions in leucine codons were used; for 2nd codon position all changes were included, and for 3rd codon position transversions were used (61). At the nucleotide level the NJ analysis was performed applying the HKY model of sequence evolution (62), whereas for amino acid sequences the Dayhoff matrix was used. For ML/QP, the TN model (63) for nucleotide sequences or the mtREV-22 model (42) for amino acid sequences were applied. Due to computational constraints bootstrap replications were limited to 100 in the case of NJ and MP analyses of amino acids, in other instances 1000 bootstrap replicates were performed. Dash (—) denotes unresolved relationship, and values in parentheses show the support for the separation between myomorph rodents and eutherians when the position of the guinea pig and the rabbit deviated from that shown in Fig. 3.
An extended ML analysis at the amino acid level was performed to investigate the support for the Marsupionta topology with different combinations of eutherians and outgroup species included. The approach made it possible to study the effect on the phylogenetic reconstruction of outgroup species deviating significantly from the mean amino acid composition of the data set.
Table 2 shows the result of an ML analysis based on amino acid sequences of nine mammals, six of which constitute eutherians included in a previous study of mammalian relationships (13). When the frog was the only outgroup, the log likelihood value for the Theria hypothesis became 2.4 standard deviations inferior to the Marsupionta topology and could thus be rejected above the 95% confidence interval. A topology with monotremes and eutherians as sister groups (an unnamed topology that has never been seriously considered) was 1.4 standard deviations inferior to the Marsupionta tree. The unnamed topology that is not rejectable with statistical confidence at the 95% level appears nevertheless highly improbable. When the chicken was used as the only outgroup the unnamed topology became virtually indistinguishable from the best topology. The Theria hypothesis still received least support, 1 standard deviation inferior to the Marsupionta tree, but could no longer be rejected with statistical significance. The findings indicate that the use of an outgroup which significantly deviates from the mean amino acid composition reduces the potential of resolution in the ML tree reconstruction. When the frog and the chicken were used together as outgroup, the support for the Marsupionta hypothesis increased slightly, although it was still impossible to distinguish between the two best alternatives with any statistical confidence.
Table 2.
Support for three possible topologies based on ML analysis of the concatenated amino acid sequences of the 12 H-strand encoded mitochondrial protein-coding genes
|
Xenopus
|
Chicken
|
Xenopus plus chicken
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Δln L | σ | pBoot | Δln L | σ | pBoot | Δln L | σ | pBoot | |
| ML analysis of 9 mammalian taxa and different outgroups* | |||||||||
| Marsupionta | 〈−31,284〉 | 93.1 | 〈−31,411〉 | 51.5 | 〈−32,536〉 | 75.9 | |||
| Theria | −47.0 | 20.0 | 0.2 | −18.2 | 16.7 | 5.3 | −26.8 | 18.6 | 4.5 |
| Unnamed | −30.9 | 21.8 | 6.7 | −2.5 | 18.7 | 43.2 | −15.8 | 19.8 | 19.6 |
| ML analysis of 23 mammalian taxa and different outgroups† | |||||||||
| Marsupionta | 〈−44,981〉 | 92.1 | 〈−45,107〉 | 52.4 | 〈−48,370〉 | 69.2 | |||
| Theria | −36.7 | 19.1 | 2.0 | −14.3 | 16.0 | 9.2 | −27.5 | 16.5 | 1.9 |
| Unnamed | −28.5 | 20.2 | 5.9 | −2.4 | 17.6 | 38.4 | −10.1 | 18.9 | 28.9 |
The likelihood value of the best topology is shown in angle brackets. The differences of log-likelihood values of alternative trees relative to that of the best tree (Δln L) as well as the standard deviations (σ) are shown (64). pBoot indicates the estimated bootstrap probability (65) among the three alternatives.
Mammals included platypus, opossum, wallaroo, rat, mouse, Caucasian human, harbor seal, cow, and whale.
Mammalian taxa as in Fig. 3.
Similar results were obtained when the data set was extended to include 24 mammalian sequences and different outgroup combinations (Table 2). Again, with only the frog as outgroup, the Theria topology could be significantly rejected. With only the chicken as outgroup the unnamed topology was again virtually indistinguishable from the best tree (Marsupionta topology), and the use of the frog and the chicken as outgroup did not significantly improve the resolution.
Analyses of mtDNA data sets have established myomorph rodents as outgroup to primates and ferungulates (14, 24). This relationship, however, is less strongly supported after the inclusion of the guinea pig and the rabbit. Consistent with a previously published phylogeny (27), the guinea pig does not form a monophyletic group with the myomorph rodents. The position of the guinea pig, however, is not entirely stable because in some analyses it joins the myomorph rodents or the rabbit, albeit with limited support. In Fig. 3 the branches marked with an asterisk (∗) were unstable and not consistently recognized by all types of data sets or all methods of phylogenetic reconstruction. Although the myomorph rodents are at the base of the eutherian tree in Fig. 3, it should be observed that the position of the Lipotyphla (hedgehog) is basal to that of myomorph rodents (24, 27). The hedgehog was not included in the data set, however, because of the deviating composition of its mtDNA sequence, but a separate ML reconstruction including all available vertebrate sequences placed the hedgehog basal to other eutherians (data not shown), and the support for the Marsupionta hypothesis remained strong (QP support value, 98.8%).
NJ of amino acid distances (based on the Dayhoff model) showed strong affiliation (94% bootstrap value) between the rabbit and the ferungulates (carnivores, perissodactyls, artiodactyls, cetaceans). Also all reconstructions based on the QP algorithm resulted in a rabbit/ferungulate relationship with support values reaching 85%. This topology is inconsistent with the reported affiliation between lagomorphs and primates (66). In the present data set a lagomorph/primate relationship was not identified by any method of tree reconstruction and occurred only at very low frequency in the bootstrap analysis. The support for the rabbit as a sister group to ferungulates is not statistically significant, however, so more extensive data will be required to conclusively answer this question. The present data set reconfirmed and corroborated the recently proposed (33) sister-group relationship between perissodactyls and carnivores.
DATING OF EVOLUTIONARY DIVERGENCES
The establishment of correct phylogenetic relationships, the calibration of differences in evolutionary rates, and the selection of a reliable reference are the essentials of molecular datings of evolutionary divergences. Considering the importance of reliable references, it is surprising that the selection of such references has been an entirely subordinate topic in molecular phylogenetics, despite the obvious fact that all calculations of evolutionary divergences will unconditionally be a direct reflection of the dating allocated to the reference. This particular topic was addressed recently in a study in which a newly established reference, the evolutionary divergence between artiodactyls and cetaceans set at 60 million years before present (MYBP), A/C-60 (67), was applied to calculate hominoid divergences (68). This application of the A/C-60 reference showed that hominoid divergences were much older than commonly acknowledged on the basis of inferential ages of local primate references.
In the present study we have applied A/C-60 for calculating several ancient evolutionary divergences among mammals. The dating of some of these divergences was recently addressed by applying as a reference the split between birds (diapsid reptiles) and mammals (synapsid reptiles) set at 310 MYBP (69). Superficially, it might appear advantageous to apply deep divergences of this kind as references for dating more recent evolutionary splits because possible errors in the dating of such references would become reduced in the dating of the more recent divergences. The efficiency of such an approach is illusory, however, because too distant references will not permit proper resolution of recent divergences. Thus the diapsid/synapsid reference did not permit resolution of the relationship among artiodactyls, rodents, and primates that has been resolved conclusively in both the present as well as previous studies (13, 14, 33, 68). A problem similar to the diapsid/synapsid reference was encountered in a study in which the chicken and the frog were used as outgroup in an effort to resolve eutherian phylogeny on the basis of all protein-coding genes of complete sequences of various mitochondrial genomes (70). In that study a (outgroup (primate (rodent, ferungulate))) relationship was identified as the most probable eutherian relationship. The identification of the correct topology was thus impaired, probably because of the combined effects of a large distance of the outgroup and different amino acid composition between some of the outgroup species and the eutherians.
The relationship between monotremes, marsupials, and eutherians has been examined on the basis of sequence data of three nuclear encoded genes (α and β hemoglobin and myoglobin). Each of the three set of data (141, 146, and 153 aa, respectively) provided support for each of the three possible topologies. The support for any particular topology was in no instance statistically significant (M. Hasegawa, personal communication).
After establishing the phylogeny shown in Fig. 3, the datings of various divergences were determined by applying A/C-60 (Table 3). Distances were estimated from amino acid sequences applying the ML and mtREV-22 model as well as 2nd codon position distances. Although 2nd codon positions contain less phylogenetic information than amino acids, they represent the most conservative part of any protein-coding data set. The two sets of datings are consistent and not separated with statistical significance. According to the amino acid data set the divergence between monotremes/marsupials and eutherians took place 130 (2nd position 143) MYBP. The divergence within the Marsupionta—i.e., between monotremes and marsupials—took place 116 (126) MYBP, and that between Australian and South American marsupials 75 MYBP. The divergence between myomorph rodents and the remaining eutherians was dated at ≈115 (125) MYBP, and that between the guinea pig and the remaining eutherians at ≈100 MYBP. The divergence between primates and ferungulates (14, 68) was dated at ≈95 MYBP. The relationships within ferungulates (33) and primates (68) have been dealt with recently and will not be repeated here.
Table 3.
Dating of ancient mammalian divergences
| Lineage | MYBP
|
|
|---|---|---|
| Amino acid | 2nd codon position | |
| Marsupials plus monotremes, eutherians | 130 ± 9.7 | 143 ± 16.8 |
| Marsupials, montremes | 116 ± 9.0 | 126 ± 15.9 |
| Myomorph rodents, remaining eutherians | 115 ± 9.0 | 125 ± 15.6 |
| Guinea pig, remaining eutherians | 98 ± 7.9 | 103 ± 13.4 |
| Primates, ferungulates | 95 ± 7.3 | 101 ± 12.2 |
| Wallaroo, opossum | 75 ± 7.1 | 76 ± 11.8 |
Datings were based on ML distances of amino acids according to the mtREV-22 matrix, and ML distances of the 2nd codon positions based on the TN model of sequence evolution (63) with the parameters of the model estimated from the data set by the puzzle program (transition/transversion parameter = 1.60 and purine transition/pyrimidine transition parameter = 1.66). As a calibration point, the artiodactyl/cetacean divergence set at 60 MYBP (A/C-60) was applied. The 95% confidence intervals were estimated according to Possion expectations of numbers of amino acid or nucleotide substitutions. The phylogenetic analysis generally joined the rabbit and the ferungulates. With that topology the dating of the divergence between the rabbit and the ferungulates is tentatively given at ≈90 MYBP.
CONCLUSIONS
The present analysis supports, in many instances significantly, a sister-group relationship between marsupials and monotremes, known as the Marsupionta hypothesis. The support provided by different molecular data sets, analytical methods, and taxon combinations in favor of a monotreme/marsupial relationship relative to a marsupial/eutherian relationship, strongly challenges the traditional concept of mammalian evolution—i.e., that the primary mammalian evolutionary distinction is that between monotremes and a common marsupial/eutherian lineage.
The presently proposed datings of mammalian divergences were based on a newly established reference, the evolutionary separation of Artiodactyla and Cetacea dated molecularly at 60 MYBP (67). The A/C-60 reference has been applied recently to provide datings for several evolutionary divergences among ferungulates (32, 33) and hominoids (68). This reference is positioned approximately midway in the time span of eutherian evolution, making it possible to approach both ancient and recent mammalian divergences without the problems associated with too distantly related nonmammalian references. As shown by the application of A/C-60, the origin of many, probably most, mammalian lineages goes back well into Cretaceous times.
Acknowledgments
We express our gratitude to Drs. Bengt Döken (Kolmården Zoo) for tissue samples, Marion Dörner who informed us on her findings on cDNA sequences of marsupial mitochondrial tRNA-Lys, and A. v. Haeseler and K. Strimmer for making the latest puzzle program (beta version 2.5) available for testing and for giving helpful hints to the program. The work was supported by the Swedish Natural Sciences Research Council and by contract ERBCHR XCT 930254 from the European Commission.
Footnotes
Abbreviations: mt, mitochondrial; MYBP, million years before present; ML, maximum likelihood; MP, maximum parsimony; NJ, neighbor joining; QP, quartet puzzling.
References
- 1.Marshall L. Zool J Linn Soc (London) 1979;66:369–410. [Google Scholar]
- 2.Carroll R L. Vertebrate Paleontolgy and Evolution. New York: Freeman; 1988. [Google Scholar]
- 3.Kermack D M, Kermack K A. The Evolution of Mammalian Characters. London: Croom Helm; 1984. [Google Scholar]
- 4.Kielan-Jaworowska Z, Bown T M, Lillegraven J A. In: Mesozoic Mammals: The First Two-Thirds of Mammalian History. Lillegraven J A, Kielan-Jaworowska Z, Clemens W A, editors. Berkeley: Univ. of California Press; 1979. pp. 221–258. [Google Scholar]
- 5.Gregory W K. Am Mus Nat Hist Bull. 1947;88:1–52. [Google Scholar]
- 6.Kühne W G. Z Morphol Tiere. 1973;75:59–64. [Google Scholar]
- 7.Kühne W G. Colloq Int CNRS. 1975;218:585–590. [Google Scholar]
- 8.Clemens W A. In: Mesozoic Mammals: The First Two-Thirds of Mammalian History. Lillegraven J A, Kielan-Jaworowska Z, Clemens W A, editors. Berkeley: Univ. of California Press; 1979. pp. 309–311. [Google Scholar]
- 9.Novacek M J, Wyss A R. Cladistics. 1986;2:257–287. doi: 10.1111/j.1096-0031.1986.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 10.Goodman M, Czelusniak J, Beeber J E. Cladistics. 1985;1:171–185. doi: 10.1111/j.1096-0031.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Retief J D, Winkfein R J, Dixon G H. Eur J Biochem. 1993;218:457–461. doi: 10.1111/j.1432-1033.1993.tb18396.x. [DOI] [PubMed] [Google Scholar]
- 12.Gemmell N J, Westerman M. J Mamm Evol. 1994;2:3–23. [Google Scholar]
- 13.Janke A, Gemmell N J, Feldmaier-Fuchs G, von Haeseler A, Pääbo S. J Mol Evol. 1996;42:153–159. doi: 10.1007/BF02198841. [DOI] [PubMed] [Google Scholar]
- 14.Janke A, Feldmaier-Fuchs G, Thomas W K, von Haeseler A, Pääbo S. Genetics. 1994;137:243–256. doi: 10.1093/genetics/137.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janke A, Pääbo S. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mörl M, Dörner M, Pääbo S. Nucleic Acids Res. 1995;23:3380–3384. doi: 10.1093/nar/23.17.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnason U, Gullberg A, Widegren B. J Mol Evol. 1991;33:556–568. doi: 10.1007/BF02102808. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fitsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Sanger F. Science. 1981;214:1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- 20.Chang Y S, Huang F L, Lo T B. J Mol Evol. 1994;38:138–155. doi: 10.1007/BF00166161. [DOI] [PubMed] [Google Scholar]
- 21.Tzeng C-S, Hui C-F, Shen S-C, Huang P C. Nucleic Acids Res. 1992;20:4853–4858. doi: 10.1093/nar/20.18.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roe B A, Ma D-P, Wilson R K, Wong J F-H. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 23.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 24.Krettek A, Gullberg A, Arnason U. J Mol Evol. 1995;41:952–957. doi: 10.1007/BF00173175. [DOI] [PubMed] [Google Scholar]
- 25.Bibb M J, Van Etten R A, Wright C T, Walberg M W, Clayton D A. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 26.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sibisa E, Saccone C. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 27.D’Erchia A M, Gissi C, Pesole G, Saccone C, Arnason U. Nature (London) 1996;381:597–599. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- 28.Lopez J V, Cevario S, O’Brien S J. Genomics. 1996;33:229–246. doi: 10.1006/geno.1996.0188. [DOI] [PubMed] [Google Scholar]
- 29.Arnason U, Gullberg A, Johnsson E, Ledje C. J Mol Evol. 1993;37:323– 330. doi: 10.1007/BF00178862. [DOI] [PubMed] [Google Scholar]
- 30.Arnason U, Johnsson E. J Mol Evol. 1992;34:493–505. doi: 10.1007/BF00160463. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, Arnason U. Gene. 1994;148:357–362. doi: 10.1016/0378-1119(94)90713-7. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Gullberg A, Arnason U. J Mol Evol. 1996;43:438–446. doi: 10.1007/BF02337515. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, Janke A, Arnason U. Mol Biol Evol. 1996;13:1167–1173. doi: 10.1093/oxfordjournals.molbev.a025681. [DOI] [PubMed] [Google Scholar]
- 34.Anderson S, de Brujin M H L, Coulson A R, Eperon I C, Sanger F, Young G. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 35.Arnason U, Gullberg A. J Mol Evol. 1993;37:312–322. doi: 10.1007/BF00178861. [DOI] [PubMed] [Google Scholar]
- 36.Arnason U, Gullberg A, Xu X. Hereditas. 1996;124:185–189. [Google Scholar]
- 37.Xu X, Arnason U. J Mol Evol. 1996;43:431–437. doi: 10.1007/BF02337514. [DOI] [PubMed] [Google Scholar]
- 38.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Arnason U. Mol Biol Evol. 1996;13:691–698. doi: 10.1093/oxfordjournals.molbev.a025630. [DOI] [PubMed] [Google Scholar]
- 40.Arnason U, Xu X, Gullberg A. J Mol Evol. 1996;42:145–152. doi: 10.1007/BF02198840. [DOI] [PubMed] [Google Scholar]
- 41.Felsenstein, J. (1991) Phylogenetic Inference Programs (phylip) (Univ. of Washington, Seattle/Univ. Herbarium/Univ. of California, Berkeley).
- 42.Adachi, J. (1995) Ph.D. Thesis (School of Mathematical and Physical Science, Tokyo).
- 43.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 44.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Fitch W M. Syst Zool. 1971;20:406–416. [Google Scholar]
- 46.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 47.Fearnley I M, Walker J E. Biochemistry. 1987;26:8247–8251. doi: 10.1021/bi00399a034. [DOI] [PubMed] [Google Scholar]
- 48.Ojala D, Montoya J, Attardi G. Nature (London) 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 49.Bennet J L, Clayton D A. Mol Cell Biol. 1990;10:2191–2201. doi: 10.1128/mcb.10.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walberg M W, Clayton D A. Nucleic Acids Res. 1981;9:5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gemmell N J, Western P S, Watson J M, Marshall Graves J A. Mol Biol Evol. 1996;13:798–808. doi: 10.1093/oxfordjournals.molbev.a025640. [DOI] [PubMed] [Google Scholar]
- 52.Yokogawa T, Watanabe Y-I, Kumazawa Y, Ueda T, Hirao I, Miura K-I, Watanabe K. Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Börner G V, Mörl M, Janke A, Pääbo S. EMBO J. 1996;15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- 54.Wolstenholme D R. Int Rev Cytol. 1992;141:173–215. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 55.Steinberg S, Cedergren R. Struct Biol. 1994;1:507–510. doi: 10.1038/nsb0894-507. [DOI] [PubMed] [Google Scholar]
- 56.Lonergan K M, Gray M W. Science. 1993;259:812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- 57.Yokobori S-I, Pääbo S. Nature (London) 1995;377:490. doi: 10.1038/377490a0. [DOI] [PubMed] [Google Scholar]
- 58.Kiss T, Fillipowicz W. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 59.Adachi J, Hasegawa M. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- 60.von Haeseler A, Janke A, Pääbo S. Verh Dtsch Zool Ges. 1993;86:119– 129. [Google Scholar]
- 61.Irwin D M, Kocher T D, Wilson A C. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 62.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 63.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 64.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 65.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;31:153–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 66.Graur D, Duret L, Gouy M. Nature (London) 1996;379:333–335. doi: 10.1038/379333a0. [DOI] [PubMed] [Google Scholar]
- 67.Arnason U, Gullberg A. Mol Biol Evol. 1996;13:407–417. doi: 10.1093/oxfordjournals.molbev.a025599. [DOI] [PubMed] [Google Scholar]
- 68.Arnason U, Gullberg A, Janke A, Xu X. J Mol Evol. 1996;43:650–661. doi: 10.1007/BF02202113. [DOI] [PubMed] [Google Scholar]
- 69.Hedges S B, Parker P H, Sibley C G, Kumar S. Nature (London) 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- 70.Adachi J, Cao Y, Hasegawa M. J Mol Evol. 1993;36:270–281. doi: 10.1007/BF00160483. [DOI] [PubMed] [Google Scholar]