Overexpression, purification and crystallization of the YjgF/YER057c/UK114-family protein from S. tokodaii strain 7 allowed the collection of a complete data set to 2.0 Å resolution.
Keywords: YjgF, YER057c, UK114, Sulfolobus tokodaii strain 7, structural genomics
Abstract
ST0811 from Sulfolobus tokodaii strain 7, a member of the YjgF/YER057c/UK114 protein family, was crystallized by the sitting-drop vapour-diffusion method using PEG 10 000 as precipitant. The crystals diffracted X-rays to beyond 2.0 Å resolution using an in-house rotating-anode generator. The crystals belonged to the rhombohedral space group R3, with hexagonal unit-cell parameters a = b = 55.0, c = 223.2 Å. The crystals contained two molecules in the asymmetric unit (V M = 2.3 Å3 Da−1) and had a solvent content of 47%.
1. Introduction
The YjgF/YER057c/UK114 family, named after the representative proteins in Escherichia coli (YjgF), yeast (YER057c) and goat (UK114), is a widely distributed family of proteins involved in a variety of biological processes. All the members are about 130 residues in length and do not occur as domains of larger proteins (Parsons et al., 2003 ▶). In bacteria and yeast, these proteins are involved in the biosynthesis of isoleucine, thiamine and purine (Enos-Berlage et al., 1998 ▶; Rappu et al., 1999 ▶). In contrast, in mammals their functions include cell-free translation-inhibitory activity and ribonuclease activity towards single-stranded RNA (Oka et al., 1995 ▶; Morishita et al., 1999 ▶). YjgF/YER057c/UK114-family members are also found in the archaea. ST0811 is a 125-residue protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7 and one of the several archaeal YjgF/YER057c/UK114-family members, although its function remains unknown. In order to predict the function of the archaeal protein ST0811 based on its molecular structure, we are analyzing the three-dimensional structure of the protein. Although the structures of bacterial orthologues such as E. coli YjgF (Volz, 1999 ▶) and Bacillus subtilis YabJ (Sinha et al., 1999 ▶) have already been solved and shown to be similar to the structure of chorismate mutase (Chook et al., 1993 ▶), to date no crystallization or crystal structure determination has been published for archaeal proteins belonging to this family. Here, we describe the crystallization and preliminary X-ray analysis of ST0811, which is 46% identical to YjgF, 40% identical to YER057c and 42% identical to UK114 in amino-acid sequence.
2. Methods and results
2.1. Purification and crystallization
Recombinant ST0811 was overexpressed in E. coli Rosetta(DE3) using the pET system (Novagen). The cell lysate was incubated at 353 K for 20 min and then centrifuged. The supernatant was applied onto a Ni–NTA agarose column (Qiagen) and the His6-tagged ST0811 was eluted with 20 mM sodium phosphate pH 7.1, 0.2 M imidazole, 0.3 M NaCl and 20 mM 2-mercaptoethanol. The His6 tag was removed by incubating His6-tagged ST0811 with thrombin (Amersham Biosciences; 10 units of enzyme per milligram of substrate protein) at room temperature for 20 h in 20 mM Tris–HCl pH 7.1. After a final purification by gel filtration using a Superdex 75 HR 10/30 column equilibrated with 20 mM Tris–HCl pH 7.1 and 0.15 M NaCl, the protein solution was dialyzed against 10 mM Tris–HCl pH 7.1 and concentrated to 12 mg ml−1 using an Apollo ultrafiltration concentrator (Orbital Biosciences). Dynamic light-scattering analysis of purified ST0811 with a Dynapro-MSTC instrument (Protein Solutions Inc.) gave a monomodal distribution and a very low polydispersity (4.9%), indicating that the sample was homogeneous and suitable for crystallization. Crystallization trials were performed by the sitting-drop vapour-diffusion method using the crystallization screening kit Crystal Screen HT (Hampton Research).
Crystals appeared in the presence of polyethylene glycol (PEG) as precipitant. After refinement of the crystallization conditions, crystals suitable for X-ray analysis were obtained in 3 d by mixing 3.0 µl protein solution and 3.0 µl reservoir solution containing 16%(w/v) PEG 10 000, 0.1 M bis-tris pH 5.3 and 0.1 M ammonium acetate. Drops were equilibrated against 0.5 ml reservoir solution at 293 K. Fig. 1 ▶ shows a typical crystal (an equilateral triangle with a side length of 0.25 mm and a depth of 0.02 mm).
Figure 1.
A crystal of ST0811 grown at 293 K using PEG 10 000 as precipitant. This crystal had a side length of 0.25 mm and a depth of 0.02 mm.
2.2. X-ray data collection and processing
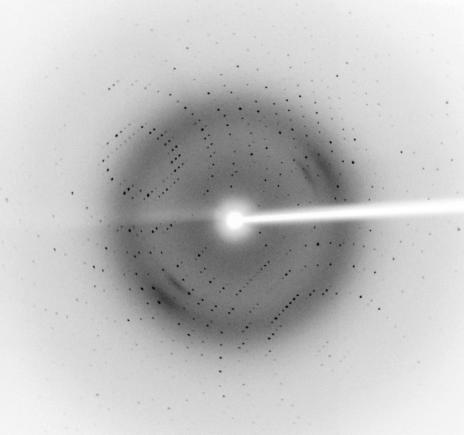
A crystal of ST0811 was picked up in a nylon loop (Hampton Research), transferred to a cryoprotectant solution containing 20%(v/v) glycerol, 18%(w/v) PEG 10 000, 0.12 M ammonium acetate and 0.12 M bis-tris buffer pH 5.3 and mounted for flash-cooling at 100 K using a GN-12 cryostat (Iwatani, Japan). Diffraction data were measured in-house using an R-AXIS VII image-plate detector mounted on an FR-E rotating-anode X-ray generator (Rigaku, Japan). X-ray data were collected using 0.5° oscillations with a crystal-to-detector distance of 150 mm. The wavelength was set to 1.542 Å and the crystals diffracted X-rays to beyond 2.0 Å resolution (Fig. 2 ▶). The diffraction images were integrated using the program MOSFLM (Leslie, 1992 ▶) and scaled using the program SCALA (Evans, 1993 ▶). The crystal belonged to the rhombohedral space group R3, with hexagonal unit-cell parameters a = b = 55.0, c = 223.2 Å. The crystal contained two molecules per asymmetric unit, according to the crystal volume per protein weight of 2.3 Å3 Da−1, with a solvent content of ∼47% (Matthews, 1968 ▶). The data statistics are shown in Table 1 ▶.
Figure 2.
An X-ray diffraction image (0.5° oscillation) from a ST0811 crystal. The edge of the detector corresponds to a resolution of 2.0 Å.
Table 1. Crystal parameters of ST0811.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.542 |
| Space group | R3 |
| Unit-cell parameters (Å) | a = b = 55.0, c = 221.6 |
| Resolution range (Å) | 22.0–2.00 (2.11–2.00) |
| Unique reflections | 16882 |
| Redundancy | 6.2 (5.9) |
| Data completeness (%) | 99.8 (99.8) |
| Rsym† | 0.055 (0.137) |
| 〈I〉/〈σ(I)〉 | 9.8 (5.1) |
R
sym = 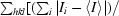
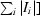 , where I
i is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I〉 is its average.
, where I
i is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈I〉 is its average.
Structure determination by molecular replacement using the coordinates of B. subtilis YabJ (50% sequence identity to ST0811; PDB code 1qd9; Sinha et al., 1999 ▶) as a search model is currently under way.
Acknowledgments
We would like to thank Dr Yutaka Kawarabayasi (Institute of Advanced Industrial Science and Technology, Japan) for providing the ST0811 gene. This research was supported in part by the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Chook, Y. M., Ke, H. & Lipscomb, W. N. (1993). Proc. Natl Acad. Sci. USA, 90, 8600–8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos-Berlage, J. L., Langendorf, M. J. & Downs, D. M. (1998). J. Bacteriol.180, 6519–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, P. R. (1993). Proceeding of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 114–122. Warrington: Daresbury Laboratory.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Morishita, R., Kawagoshi, A., Sawasaki, T., Madin, K., Ogasawara, T., Oka, T. & Endo, Y. (1999). J. Biol. Chem.274, 20688–20692. [DOI] [PubMed] [Google Scholar]
- Oka, T., Tsuji, H., Noda, C., Sakai, K., Hong, Y. M., Suzuki, I., Muñoz, S. & Natori, Y. (1995). J. Biol. Chem.270, 30060–30067. [DOI] [PubMed] [Google Scholar]
- Parsons, L., Bonander, N., Eisenstein, E., Gilson, M., Kairys, V. & Orban, J. (2003). Biochemistry, 42, 80–89. [DOI] [PubMed] [Google Scholar]
- Rappu, P., Shin, B. S., Zalkin, H. & Mäntsälä, P. (1999). J. Bacteriol.181, 3810–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S., Rappu, P., Lange, S. C., Mantsala, P., Zalkin, H. & Smith, J. L. (1999). Proc. Natl Acad. Sci. USA, 96, 13074–13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz, K. (1999). Protein Sci.8, 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]