Crystals of P. platycephala chintinase/lectin (PPL-2) belong to the orthorhombic space group P212121, with unit-cell parameters a = 55.19, b = 59.95, c = 76.60 Å. The preliminary cystal structure of PPL-2 was solved at a resolution of 1.73 Å by molecular replacement, presenting a correlation coefficient of 0.558 and an R factor of 0.439.
Keywords: chitin-binding proteins, chitinases, Parkia platycephala, lectins
Abstract
A chitin-binding protein named PPL-2 was purified from Parkia platycephala seeds and crystallized. Crystals belong to the orthorhombic space group P212121, with unit-cell parameters a = 55.19, b = 59.95, c = 76.60 Å, and grew over several days at 293 K using the hanging-drop method. Using synchrotron radiation, a complete structural data set was collected to 1.73 Å resolution. The preliminary crystal structure of PPL-2, determined by molecular replacement, presents a correlation coefficient of 0.558 and an R factor of 0.439. Crystallographic refinement is in progress.
1. Introduction
Chitin, a natural homopolymer composed of β(1–4)-linked N-acetylglucosamine (GlcNAc)n, is a major component of the exoskeleton of fungi (comprising up to 30% of fungal cell walls) and invertebrates. It is easily obtained from marine invertebrates, insects and algae (Patil et al., 2000 ▶).
The complete enzymatic hydrolysis of chitin to free N-acetylglucosamine residues is performed by a chitinolic system and is known to be a continuous reaction. Different organisms produce a wide variety of hydrolytic enzymes that exhibit different substrate specificities. Some of them are called chitinases, which are enzymes that catalyze the hydrolysis of chitin. These proteins are a large and diverse group of enzymes that differ in their molecular structure, substrate specificity and catalytic mechanism (Kasprzewska, 2003 ▶). Specificity for chitin oligosaccharide is not a feature that is exclusive to the chitinases. Proteins named ‘chitin-binding lectins’ or ‘hevein-like lectins’ also possess affinity for N-acetylglucosamine residues, but cannot catalyze the hydrolysis of chitin (Van Damme et al., 1998 ▶).
Several chitinases have been found in plants (angiosperms and gymnosperms) and are present in diverse tissues. Most are expressed by stress factors such as infection. Plants use chitinases as a defence against pathogenic fungi, but the enzymes may also perform other functions (Peumans et al., 2002 ▶). Some chitinases have industrial and agricultural applications, such as in the biocontrol of pathogenic fungi and insects, as a target for biopesticides and in the production of chitooligosaccharides (Kasprzewska, 2003 ▶; Patil et al., 2000 ▶).
Plant lectins with chitinase activity are poorly described in the literature. The acidic chitinase from Brassica juncea shows a structure that is distinct from those observed for chitinases studied previously. This difference is characterized by the presence of two chitin-binding sites (Zhao & Chye, 1999 ▶), which permit this protein to agglutinate cells and may provide an advantage over other chitinases in antimicrobial and antifungal activity (Chye et al., 2005 ▶).
Many carbohydrate-binding proteins have been reported, in particular those purified from plants (Moreno et al., 2004 ▶; Gadelha et al., 2005 ▶). The majority are from the Leguminosae family and comprise lectins and chitinases from diverse sources. Legume lectins have been well studied as a model of carbohydrate recognition. In the subfamily Mimosoideae, however, apart from Parkia plathycephala 2 (PPL-2), only the seed lectins from P. speciosa (Suvachittanont & Peutpaiboon, 1992 ▶), P. javanica (Utarabhand & Akkayanont, 1995 ▶), P. platycephala (Cavada et al., 1997 ▶) and P. discolor (Cavada et al., 2000 ▶) have been isolated and characterized in detail. Moreover, crystal structures are only available for P. platycephala lectin (PPL-1) in native form (PDB code 1zgr) and in complex with 5-bromo-4-chloro-3-indolyl-α-d-mannose (PDB code 1zgs).
Mass-spectrometric analysis indicates that the PPL-2 monomer is not glycosylated and contains six cysteine residues that are involved in three disulfide bonds; PPL-2 gives a main mass peak at 29 407. Functional analysis reveals that PPL-2 recognizes carbohydrates on red blood cells and agglutinates trypsin-treated rabbit erythrocytes (128 haemagglutinating units per millilitre). In addition, PPL-2 can hydrolyze β(1–4)-glycosidic linkages between 2-acetoamido-2-deoxy-β-d-glucopyranoses present in chitin. The exact mechanism of glycoside hydrolysis has been described by Cavada et al. (2005 ▶) and this mechanism reveals an endochitinase activity to be associated with PPL-2 from the elution times found for the GlcNAc, (GlcNAc)2 and (GlcNAc)3 standards. Hence, PPL-2 is the first and is a remarkable chimerolectin from the Mimosoideae, with the dual property of hydrolyzing chitin and binding sugar moieties on red blood cells (Cavada et al., 2005 ▶).
In order to establish the crystal structure of this new member of the chitin-binding proteins, this work reports the crystallization and preliminary X-ray diffraction analysis of a hevamine-like protein from P. platycephala seeds, named PPL-2, that has the ability to agglutinate cells and shows inhibitory effects in the growth of bacterial colonies and nematode-egg eclosion (Castellón, 2004 ▶; Cavada et al., 2005 ▶).
2. Material and methods
2.1. Purification and crystallization
Soluble proteins were extracted from the seeds of P. platycephala Benth in an extraction solution (0.1 M HCl with 0.1 M NaCl). After centrifugation, the supernatant was neutralized with sodium hydroxide (NaOH) and the neutralized solution was submitted to precipitation with ammonium sulfate. The fraction 0/60 was resuspended in 0.05 M Tris–HCl buffer with 0.1 M NaCl pH 7.0 and exhaustively dialyzed against this buffer. The protein was purified by affinity chromatography on a Red-Sepharose CL-6B (23.0 × 2.5 cm) column equilibrated with the same buffer; elution of the non-interacting material took place using the equilibration buffer and the protein was eluted with 0.05 M Tris–HCl with 3.0 M NaCl pH 7.0 and finally dialyzed against Milli-Q water and lyophilized (Castellón, 2004 ▶; Cavada et al., 2005 ▶).
For crystallization trials, the purified lectin was dissolved at a concentration of 7.5 mg ml−1 in Milli-Q water. Microcrystals of PPL-2 were grown in Linbro plates at 293 K by the vapour-diffusion/sparse-matrix method (Jancarik & Kim, 1991 ▶) in hanging drops using Crystal Screen from Hampton Research. The drops were composed of equal volumes (3 µl) of protein solution and reservoir solution [0.2 M ammonium acetate, 0.1 M trisodium citrate dehydrate pH 5.6 and 30%(w/v) polyethylene glycol 4000] and were equilibrated against 500 µl reservoir solution. Microcrystals were seeded into a new drop containing the same crystallization solution and an equal protein volume.
2.2. X-ray data collection
X-ray diffraction data were collected at a wavelength of 1.4727 Å using a synchrotron-radiation source (CPr station, Laboratorio Nacional de Luz Síncrotron, Campinas, Brazil) and a CCD detector (MAR Research) with a crystal-to-detector distance of 70.00 mm at a temperature of 100 K. To avoid freezing, crystals were soaked in a cryoprotectant solution containing 75% mother liquor and 25% glycerol. Using an oscillation range of 1.0° and an exposure time of 30 s per frame, 90 images were collected to a maximum resolution of 1.73 Å. Data were indexed, integrated and scaled using MOSFLM and SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
2.3. Molecular replacement
Sequence-alignment analysis was performed using programs that compared the N-terminal sequence of PPL-2 with those of all the non-redundant bank of proteins deposited in the National Center of Biotechnology Information (NCBI). Local and multiple alignments were carried out using BLAST (Altschul et al., 1990 ▶) and CLUSTALW (Thompson et al., 1994 ▶), respectively. To perform multiple alignments, plant chitinases from Nicotiana tabacum, Phytolacca americana, Glycine max, Zea mays, Vitis vinifera, Arabdopsis thaliana, Vigna unguiculata and Hevea brasiliensis were used.
The molecular-replacement method was used to determine the crystal structure of PPL-2 using the AMoRe software (Navaza, 1994 ▶). Rotation and translation functions were performed using data in the resolution range 15–3.0 Å. The best solution for each model was selected based on the magnitude of the correlation coefficient and the R factor. Four space groups were tested (P222, P21212, P2221 and P212121) using the hevamine protein coordinates (PDB code 1hvq, chain A; Terwisscha van Scheltinga et al., 1996 ▶).
3. Results and discussion
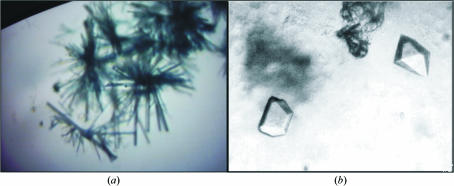
Microcrystals grew in a month using condition No. 9 of Crystal Screen from Hampton Research; when submitted to the seeding experiment, a crystal cluster was observed (Fig. 1 ▶ a) after another month. The drop was perturbed using a fine hair and after about 20 weeks suitable crystals were obtained (Fig. 1 ▶ b). The best crystals grew to approximate dimensions of 0.3 × 0.2 × 0.3 mm.
Figure 1.
PPL-2 crystals. (a) Crystal cluster from PPL-2 seeding. (b) Crystals of PPL-2 diffracted to 1.73 Å.
Crystals of PPL-2 were grown by the hanging-drop vapour-diffusion method. PPL-2 crystals belong to the orthorhombic space group P212121, with unit-cell parameters a = 55.19, b = 59.95, c = 76.70 Å. The volume of the unit cell is 253 757.48 Å3, which is compatible with one monomer in the asymmetric unit, with a V M of 2.3 Å3 Da−1 (Matthews, 1968 ▶). A summary of the data-collection statistics is given in Table 1 ▶.
Table 1. X-ray diffraction data collection.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.431 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 55.19, b = 59.95, c = 76.70 |
| Resolution range (Å) | 32.27–1.73 |
| Unique reflections | 25945 |
| Completeness (%) | 95.5 (95.5) |
| 〈I/σ(I)〉 | 13.1 (2.4) |
| Rsym (%) | 4.0 (22.8) |
| Rfull (%) | 3.6 (16.3) |
Sequence-alignment analysis permitted the retrieval of a viable search model to submit these data to molecular replacement. The N-terminal amino-acid sequence from PPL-2 (GGIVVWGQNGGEGTLTSTCESGLYQIVNIAFLSQFGGGRRV) is completely different from that found in PPL-1, a lectin isolated from seeds of P. platycephala (SLKGMISVGPWGGSGGNYWSFKANHAITEIVIHVKDNIKS; Cavada et al., 1997 ▶). Based on local alignments of PPL-2, similarity has been found with chitinases, proteins that are reported to be related to defence mechanisms in plants. These alignments show that PPL-2 exhibits a high sequence similarity to type III chitinases. One of these proteins is hevamine, a chitinase and lysozyme protein found in latex from H. brasiliensis, which has the N-terminal sequence GGIAIYWGQNGNEGTLTQTCSTRKYSYVNIAFLNKFGNGQ. Acidic chitinases extracted from the leaves of G. max (Watanabe et al., 1999 ▶), Z. mays (Didierjean et al., 1996 ▶) and A. thaliana (Kawabe et al., 1997 ▶) also show similarity with PPL-2, as can be observed in Fig. 2 ▶.
Figure 2.
Multiple alignment of the N-terminal sequence of PPL-2 with those of plant chitinases. GmCHI, acidic chitinase from G. max; ZmCHI, acidic chitinase from Z. mays; Hevamina, chitinase/lysozyme from H. brasiliensis; VvCHI, precursor of acidic chitinase from Vitis vinifera; AtCHI, acidic endochitinase from A. thaliana; VuCHI, basic endochitinase type III from Vigna unguiculata; PaCHI, chitinase-B from Phytolacca americana; NtCHI, basic endochitinase type III from N. tabacum.
The N-terminal alignment of PPL-2 and several chitinases demonstrated a degree of similarity estimated at 72% with basic endochitinase type III from N. tabacum, 71% with chitinase-B from Phytolacca americana, 71% with acidic chitinases from G. max and Z. mays, 66% with the precursor of the acidic chitinase (Precursor) from Vitis vinifera, 60% with acidic endochitinase from A. thaliana, 61% with basic endochitinase type III from Vigna unguiculata and 64% with hevamine, a chitinase/lysozyme from H. brasiliensis. Despite the high similarity observed among all the alignments, hevamine is the only one in which the polypeptidic fragment corresponds to the N-terminal region as in PPL-2 and is thus a good search model for molecular replacement. The best results were obtained for space group P212121, resulting in a correlation coefficient and R factor of 55.8 and 43.9%, respectively.
Initial crystallographic refinement was performed using rigid-body refinement followed by the maximum-likelihood method with the REFMAC5 software (Collaborative Computational Project, Number 4, 1994 ▶), resulting in a model with an R factor of 29.7% and an R free of 33.1%. Complete refinement of the structure of PPL-2 is in progress.
Acknowledgments
This work was partly financed by Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Universidade Regional do Cariri (URCA), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Laboratório Nacional de Luz Síncrotron (LNLS), Campinas, Brazil and FAPESP (SMOLBNet, 01/07532-0). BSC and WFA are senior investigators of CNPq.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol.215, 403–410. [DOI] [PubMed] [Google Scholar]
- Castellón, R. E. R. (2004). Tese de Doutorado apresentada ao Departamento de Fitotecnia, Universidade Federal do Ceará, Fortaleza-Ceará, Brazil.
- Cavada, B. S., Castellón, R. E. R., Vasconcelos, G. G., Delatorre, P., Nagano, C. S., Nascimento, K. S., Souza, E. P., Radis-Baptista, G., Leroy, Y., Toyama, M. H., Sampaio, A. H., Pinto, V. P. T. & Debray, H. (2005). Submitted.
- Cavada, B. S., Madeira, S. V. F., Calvete, J. J., Sousa, L. A. G., Bomfim, L. R., Dantas, A. R., Lopes, M. C., Grangeiro, T. B., Freitas, B. T., Pinto, V. P. T., Leite, K. B. & Ramos, M. V. (2000). Prep. Biochem. Biotech.30, 271–280. [DOI] [PubMed]
- Cavada, B. S., Santos, C. F., Grangeiro, T. B., Silva, L. I. M. M., Campos, M. J. O., Sousa, F. A. M. & Calvete, J. J. (1997). Physiol. Mol. Biol. Plants, 3, 109–115.
- Chye, M. L., Zhao, K. J., He, Z. M., Ramalingam, S. & Fung, K. L. (2005). Planta, 220, 717–730. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Didierjean, L., Frendo, P., Nasser, W., Genot, G., Marivet, J. & Burkard, G. (1996). Planta, 199, 1–8. [DOI] [PubMed] [Google Scholar]
- Gadelha, C. A. A., Moreno, F. B. M. B., Santi-Gadelha, T., Cajazeiras, J. B., Rustiguel, J. K. R., Freitas, B. T., Canduri, F., Delatorre, P. & de Azevedo, W. F. Jr (2005). Acta Cryst. F61, 87–89. [DOI] [PMC free article] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kasprzewska, A. (2003). Cell Mol. Biol. Lett.8, 809–824. [PubMed] [Google Scholar]
- Kawabe, A., Innan, H., Terauchi, R. & Miyashita, N. T. (1997). Mol. Biol. Evol.14, 1303–1315. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Moreno, F. B. M. B., Delatorre, P., Freitas, B. T., Rocha, B. A. M., Souza, E. P., Facó, F., Canduri, F., Cardoso, A. L. H., Freire, V. N., Lima-Filho, J. L., Sampaio, A. H., Calvete, J. J., de Azevedo, W. F. Jr & Cavada, B. S. (2004). Acta Cryst. D60, 1493–1495. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Patil, R. S., Ghormade, V. V. & Deshpande, M. V. (2000). Enzyme Microb. Technol.26, 473–483. [DOI] [PubMed] [Google Scholar]
- Peumans, W. J., Proost, P., Swennen, R. L. & Van Damme, E. J. (2002). Plant Physiol.130, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvachittanont, W. & Peutpaiboon, A. (1992). Phytochemistry, 31, 4065–4070.
- Terwisscha van Scheltinga, A. C., Hennig, M. & Dijkstra, B. W. (1996). J. Mol. Biol.262, 243–257. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utarabhand, P. & Akkayanont, P. (1995). Phytochemistry, 38, 281–285.
- Van Damme, J. M., Peumans, W. J., Barre, A. J. & Rougé, P. J. (1998). Crit. Rev. Plant Sci.17, 575–692.
- Watanabe, A., Nong, V. H., Zhang, D., Arahira, M., Yeboah, N. A., Udaka, K. & Fukazawa, C. (1999). Biosci. Biotechnol. Biochem.63, 251–256. [DOI] [PubMed] [Google Scholar]
- Zhao, K. J. & Chye, M. L. (1999). Plant Mol. Biol.40, 1009–1018. [DOI] [PubMed] [Google Scholar]