Recombinant β-glucosidase from wheat seedlings complexed with a substrate aglycone has been crystallized in a hexameric active form. A diffraction data set has been collected at 1.7 Å.
Keywords: β-glucosidases, DIMBOA, hexamers, wheat
Abstract
The wheat β-glucosidase TaGlu1b, which is only active in a hexameric form, was tagged with 6×His at the N-terminus, overexpressed in Escherichia coli and purified in two steps. The protein complexed with a substrate aglycone was crystallized at 293 K from a solution containing 10 mM HEPES pH 7.2, 1 M LiSO4 and 150 mM NaCl using the hanging-drop vapour-diffusion method. Diffraction data were collected to 1.7 Å at the Photon Factory. The crystal belongs to space group P4132, with unit-cell parameters a = b = c = 194.65 Å, α = β = γ = 90°. The asymmetric unit was confirmed by molecular-replacement solution to contain one monomer, giving a solvent content of 72.1%.
1. Introduction
β-Glucosidases (EC 3.2.1.21) are one of the largest groups of enzymes in the glycoside hydrolase families and belong to families 1 or 3 (Henrissat, 1991 ▶). These enzymes are responsible for cleavage of the β-glucosidic linkage between two (or more) sugars or between a sugar and an aglycone. In plants, β-glucosidases are involved in lignification (Dharmawardhana et al., 1995 ▶), regulation of the physiological activity of cytokinin (Falk & Rask, 1995 ▶; Haberer & Kieber, 2002 ▶), regulation of the biosynthesis of IAA (Ljung et al., 2001 ▶; Persans et al., 2001 ▶) and chemical defence against pathogens and herbivores (Niemeyer, 1988 ▶; Zagrobelny et al., 2004 ▶). Many secondary products in plants exist in the form of a glucoconjugate with one or two glucose units attached to their hydroxy or thiol groups. Since glucosylation changes physiological activity, chemical stability and solubility in water, β-glucosidases play important roles by cleaving glucosidic bonds to release monosaccharides and active aglycones such as cyanogenic compounds, flavonoids and hydroxamic acids. The glucosidases involved in the hydrolysis of plant secondary metabolites are members of the family 1 glycohydrolases.
In recent years, we have purified and characterized β-glucosidases from young wheat and rye (Sue et al., 2000a ▶,b ▶). Both plants accumulate hydroxamic acids (Hxs; 2,4-dihydroxy-1,4-benzoxazin-3-one, DIBOA, and its 7-methoxy derivative, DIMBOA) as defensive compounds against pathogens and herbivores (Niemeyer, 1988 ▶). These compounds are glucosylated in intact plants and are stored in different subcellular compartments from the glucosidase. Although the wheat and rye glucosidases hydrolyze DIBOA-Glc and DIMBOA-Glc efficiently, the preferred natural substrate for each glucosidase is consistent with the predominant Hx species in each plant, namely DIMBOA-Glc in wheat and DIBOA-Glc in rye. These observations raise the issue of how plants have developed these defence chemicals and the enzymes involved in their metabolism.
Recently, we cloned three β-glucosidase genes from wheat seedlings and expressed them in Escherichia coli. The recombinant glucosidases formed a hexamer, the active form, consisting of 64 kDa monomers (manuscript in preparation). The hexamer was easily dissociated into monomers, resulting in loss of activity. Crystal structures of family 1 β-glucosidases have been isolated from four plant species: white clover (Barrett et al., 1995 ▶), white mustard (Burmeister et al., 1997 ▶), maize (Czjzek et al., 2001 ▶; Zouhar et al., 2001 ▶) and sorghum (Verdoucq et al., 2004 ▶). The wheat enzyme, however, differs in its active oligomeric structure, since wheat glucosidase must be hexameric to be active as a hydrolase, whereas the other plant glucosidases function as dimers. The crystal structures of family 1 β-glucosidases from several bacteria have been solved as octamers (Sanz-Aparicio et al., 1998 ▶; Hakulinen et al., 2000 ▶). The wheat enzyme, however, shows lower homologies (∼35%) with bacterial enzymes than with those of plants (∼60%). Thus, it is of interest to compare the monomer-association properties from the quaternary structures. We therefore aimed to crystallize wheat β-glucosidase as a hexamer in order to clarify the relationship between its structure and activity.
2. Materials and methods
2.1. Cloning, expression and purification
Wheat β-glucosidase (TaGlu1b) was expressed with a 6×His tag at the N-terminus in E. coli. The DNA fragment corresponding to mature TaGlu1b, with NcoI and XhoI recognition sites at the 5′ and 3′ ends, respectively, was amplified by PCR using KOD DNA polymerase (Toyobo). The resulting fragment was introduced into the NcoI and XhoI sites of pET30a and the product was transformed into BL21 CodonPlus (DE3)-RIL (Stratagene). Luria–Bertani (LB) broth (3 ml) containing 50 µg ml−1 kanamycin and chloramphenicol was inoculated with a single colony of the E. coli and was incubated at 310 K and 150 rev min−1 for 6 h. A 2 ml aliquot of the culture was then transferred to 150 ml LB broth supplemented with the same antibiotics and was further incubated at 310 K and 150 rev min−1 until the OD600 of the medium reached 0.6. Expression of TaGlu1b was induced by adding isopropyl β-thiogalactopyranoside to a final concentration of 1 mM, followed by incubation at 293 K and 150 rev min−1 for 16 h. The cells were recovered by centrifugation at 15 000g for 15 min and were resuspended in 15 ml ice-cold HEPES pH 7.2 containing 80 µl protease-inhibitor cocktail for purification of the His-tagged protein (Sigma). The cell was disrupted by six bouts of sonication (100 W) for 20 s at 1 min intervals. The lysate was then centrifuged at 15 000g for 20 min at 277 K.
The expressed TaGlu1b was purified using a HiTrap Chelating HP column (Amersham Biosciences) according to the manufacturer’s instructions. The 60–300 mM imidazole fraction was collected and concentrated by ultrafiltration. The glucosidase was further purified by gel filtration on Superdex 200 (Amersham Biosciences) equilibrated with 50 mM HEPES pH 7.2 and 150 mM NaCl. The fraction containing glucosidase activity (about 300 kDa) was pooled and concentrated by ultrafiltration. The amount of protein was estimated according to the method of Bradford (1976 ▶), using BSA as standard.
2.2. Crystallization and preliminary X-ray diffraction
The initial crystallization conditions were screened using Hampton Research Crystal Screens I and II (Jancarik & Kim, 1991 ▶); the final condition was 10 mM HEPES pH 7.2, 1 M LiSO4 and 150 mM NaCl as the reservoir buffer. All crystallization experiments were performed using the hanging-drop vapour-diffusion method in a 24-well tissue-culture VDX plate at 293 K. Drops consisting of 1 µl protein solution (3.7 mg ml−1) and 1 µl reservoir solution were used for the initial screening and drops consisting of 1.5 µl protein solution and 1.5 µl reservoir solution were used for the final condition.
To obtain the complex of TaGlu1b and its substrate aglycone, DIMBOA, crystals were soaked in the crystallization buffer with 0.5 mM DIMBOA and 30% glycerol as a cryoprotectant for 15 min and were then cooled in a nitrogen stream at 100 K. The diffraction data were processed using the HKL2000 program (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
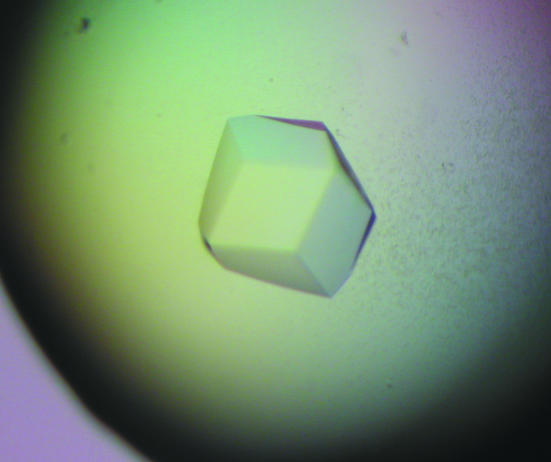
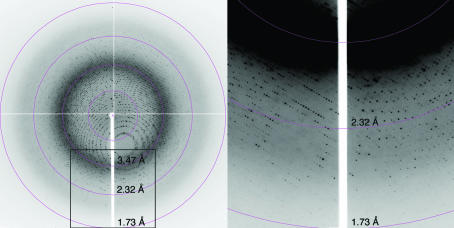
The 6×His-tagged TaGlu1b was expressed strongly in the soluble fraction in E. coli and we were able to obtain 2 mg of the purified protein from 150 ml culture. From the initial screening, crystals were obtained from both 0.1 M HEPES pH 7.5, 2 M ammonium sulfate, 2% PEG 400 and 10 mM HEPES pH 7.2, 1 M LiSO4, 150 mM NaCl. The first set of conditions produced only tiny crystals, so the latter conditions were subsequently used. The crystals were typically 0.2 mm in their longest dimension, with a maximum of around 0.3 mm (Fig. 1 ▶); we used the largest crystals for data analysis. Crystals of TaGlu1b complexed with DIMBOA were obtained by soaking and diffracted to 1.7 Å on beamline BL-6A at the Photon Factory (Fig. 2 ▶). However, crystals without the substrate diffracted to less than 2.0 Å. The mosaicity of the crystals was very low (∼0.12) and R merge was 0.079 with high redundancy (20). Table 1 ▶ shows the data-collection and related processing statistics.
Figure 1.
Crystals of TaGlu1b obtained using 10 mM HEPES pH 7.2, 1 M LiSO4, 150 mM NaCl. These crystals appeared in one week at 293 K and have a maximum size of 0.3 mm along one dimension.
Figure 2.
A representative diffraction image of a crystal of TaGlu1b complexed with DIMBOA collected on beamline BL6A at the Photon Factory using an ADSC Quantum 4 detector. The box in the left image indicates the position of the image magnified on the right.
Table 1. Data-collection and processing statistics.
Values in parentheses correspond to the outer resolution shell.
| Wavelength (Å) | 1.000 |
| Space group | P4132 |
| Unit-cell parameters | |
| a = b = c (Å) | 194.65 |
| α = β = γ (°) | 90 |
| Matthews coefficient (Å3 Da−1) | 4.45 |
| Molecules per AU | One monomer |
| Solvent content (%) | 72.1 |
| Resolution range (Å) | 50–1.70 (1.76–1.70) |
| Total observations | 2667849 |
| Unique reflections | 137005 |
| Average I/σ(I) | 62.0 (6.6) |
| Rmerge | 0.079 (0.564) |
| Completeness (%) | 99.8 (98.0) |
A molecular-replacement solution using the β-glucosidase from sorghum (PDB code 1v03; Verdoucq et al., 2004 ▶) as the search model was obtained using the program MOLREP (Vagin & Teplyakov, 1997 ▶). From this solution, the asymmetric unit was found to contain one monomer and the solvent content was 72.1%, which is relatively high despite the high resolution. Model building and refinement of this structure are under way.
Several structures of O-β-glucosidases from plants, including white clover, maize and sorghum, have been determined. The resolution obtained in the present study is better, however, and promises to provide a better understanding of the relation between the structure and activity of O-β-glucosidases.
Acknowledgments
We are grateful for access to and user support at the synchrotron facilities of the Photon Factory.
References
- Barrett, T., Suresh, C. G., Tolley, S. P., Dodson, E. J. & Hughes, M. A. (1995). Structure, 3, 951–960. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Burmeister, W. P., Cottaz, S., Driguez, H., Iori, R., Palmieri, S. & Henrissat, B. (1997). Structure, 5, 663–675. [DOI] [PubMed] [Google Scholar]
- Czjzek, M., Cicek, M., Zamboni, V., Burmeister, W. P., Bevan, D. R., Henrissat, B. & Esen, A. (2001). Biochem. J.354, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana, D. P., Ellis, B. E. & Carlson, J. E. (1995). Plant Physiol.107, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A. & Rask, L. (1995). Plant Physiol.108, 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G. & Kieber, J. J. (2002). Plant Physiol.128, 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakulinen, H., Paavilainen, S., Korpela, T. & Rouvinen, J. (2000). J. Struct. Biol.129, 69–79. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). Biochem. J.280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Ljung, K., Ostin, A., Lioussanne, L. & Sandberg, G. (2001). Plant Physiol.125, 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer, H. (1988). Phytochemistry, 11, 3349–3358.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Persans, M. W., Wang, J. & Schuler, M. A. (2001). Plant Physiol.125, 1126–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Aparicio, J., Hermoso, J. A., Martinez-Ripoll, M., Lequerica, J. L. & Polaina, J. (1998). J. Mol. Biol.275, 491–502. [DOI] [PubMed] [Google Scholar]
- Sue, M., Ishihara, A. & Iwamura, H. (2000a). Plant Sci.155, 67–74. [DOI] [PubMed] [Google Scholar]
- Sue, M., Ishihara, A. & Iwamura, H. (2000b). Planta, 210, 432–438. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Verdoucq, L., Moriniere, J., Bevan, D. R., Esen, A., Vasella, A., Henrissat, B. & Czjzek, M. (2004). J. Biol. Chem.279, 31796–31803. [DOI] [PubMed] [Google Scholar]
- Zagrobelny, M., Bak, S., Rasmussen, A. V., Jorgensen, B., Naumann, C. M. & Moller, B. L. (2004). Phytochemistry, 65, 293–306. [DOI] [PubMed] [Google Scholar]
- Zouhar, J., Vévodová, J., Marek, J., Damborský, J., Su, X.-D. & Brzobahatý, B. E. (2001). Plant Physiol.127, 973–985. [PMC free article] [PubMed] [Google Scholar]