Further understanding of the structure and function of plasma apolipoproteins requires the determination of their high-resolution structures when complexed with lipids. In these studies, the production of homogeneous, biologically active lipoprotein particles of apolipoprotein E complexed with dipalmitoylphosphatidylcholine and their crystallization and X-ray diffraction are demonstrated.
Keywords: plasma apolipoproteins, apolipoprotein E
Abstract
High-resolution structural information is available for several soluble plasma apolipoproteins (apos) in a lipid-free state. However, this information provides limited insight into structure–function relationships, as this class of proteins primarily performs its functions of lipid transport and modulation of lipid metabolism in a lipid-bound state on lipoprotein particles. Here, the possibility of generating homogeneous lipoprotein particles that could be crystallized was explored, opening the possibility of obtaining high-resolution structural information by X-ray crystallography. To test this possibility, apoE4 complexed with the phospholipid dipalmitoylphosphatidylcholine was chosen. Uniform particles containing 50% lipid and 50% apoE4 were obtained and crystallized using the hanging-drop method. Two crystal forms diffract to beyond 8 Å resolution.
1. Introduction
Apolipoprotein (apo) E serves a variety of functions in plasma cholesterol and triglyceride metabolism and transports and redistributes lipids among various cells, tissues and organs (Mahley, 1988 ▶). It interacts with members of the low-density lipoprotein (LDL) receptor family and the heparin sulfate proteoglycan/LDL receptor-related protein pathway (Mahley et al., 1994 ▶). The protein is polymorphic, with three common isoforms: apoE2, apoE3 and apoE4 (Weisgraber, 1994 ▶). ApoE also plays a key role in neurobiology and apoE4 is a major risk factor for Azheimer’s disease (Corder et al., 1993 ▶; Saunders et al., 1993 ▶; Strittmatter et al., 1993 ▶) and other forms of central nervous system stress (Mayeux et al., 1995 ▶; Roses & Saunders, 1997 ▶; Slooter et al., 1997 ▶; Teasdale et al., 1997 ▶; Fazekas et al., 2000 ▶; Drory et al., 2001 ▶).
Although X-ray structures of the amino-terminal domains of lipid-free apoE2, apoE3 and apoE4 provided considerable insight into the functions of apoE (Weisgraber, 1994 ▶), the next level of understanding and challenge requires the determation of physiologically relevant structures of apoE associated with lipid. In this study, we describe the generation of uniform biologically active lipoprotein particles of apoE4 complexed with dipalmitoylphosphatidylcholine (DPPC) and their crystallization and preliminary X-ray analysis.
2. Materials and methods
2.1. ApoE4–DPPC particle production and characterization
DPPC in chloroform (Avanti Polar Lipids 850355) was dried in a glass tube under a stream of nitrogen. A vacuum was applied to remove all traces of the chloroform before reconstitution to 20 mg ml−1 in Tris-buffered saline (TBS; 10 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.25 mM EDTA, 0.0005% NaN3). Sodium cholate (30 mg ml−1 in TBS) was added to the DPPC at a molar ratio of 1.4:1 and incubated for 1 h at 314 K, with vortexing every 10 min. The mixture was cooled to room temperature and apoE4 was added to a molar ratio of 1:46 (apoE:DPPC). After 10 min incubation at room temperature and 1 h incubation at 314 K, the cholate was removed with washed Bio-beads SM2 (Biorad 152-3920) at a ratio of 1 g Bio-beads to 2 mg cholate. The apoE–DPPC particles were purified on a Superdex 200 (Pharmacia) size-exclusion column with TBS as the running buffer and concentrated to ∼2 mg ml−1 (Lowry et al., 1951 ▶) and 1% 1,2,3-heptanetriol was added.
2.2. Negative-staining electron microscopy
DPPC particles were stained with uranyl acetate on the surface of carbon-fluid grids as described by Dong et al. (1998 ▶). Electron micrographs (200 000×) were imported with a video camera into an Image 1/AT image-analysis system. The particle size was analyzed with system software (v.4.03a, Universal Imaging Corporation). Multiple areas on a single grid were sampled.
2.3. Phospholipid analysis
The phospholipid content of the apoE4–DPPC particles was determined with a quantitative enzymatic colorimetric method and compared with known phospholipid standards (Wako Chemicals 990-54009E).
2.4. Cross-linking
20 mg ml−1 dimethyl suberimidate in 1 M triethanolamine pH 9.7 was incubated with apoE4–DPPC and control apoE4 in PBS for 90 min at room temperature and quenched with Tris–HCl pH 6.8. After vacuum drying, the samples were analyzed by SDS–PAGE.
2.5. LDL receptor-binding assay
The biological activity of the apoE4–DPPC particles was determined in a standard competitive binding assay using 125I-labelled human LDL and cultured human fibroblasts as described in Arnold et al. (1992 ▶). As a control, apoE4 was combined with dimyristoylphosphatidylcholine (DMPC) at a ratio of 1:3.75(w:w) and isolated by density-gradient ultracentrifugation (Innerarity et al., 1979 ▶).
3. Crystallization screening
Isolated apoE4–DPPC complexes (∼2 mg ml−1) were initially screened with a sparse-matrix approach using commercial Hampton Research Crystal Screens with the hanging-drop format with equal volumes of protein solution and well buffer. The results of these trials were used to develop subsequent screens, which included various molecular weights of polyethylene glycol (PEG), salts and buffers, as well as a wide variety of additives contained in Hampton Kits.
4. Data collection and processing
Data were collected at beamlines 8.3.1, 8.2.2 and 8.2.1 at the Advanced Light Source. Data were processed and scaled with XDS (Kabsch, 1993 ▶).
5. Results and discussion
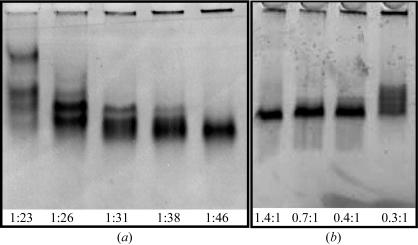
DPPC has a transition temperature of 314 K, which offers some potential advantage in handling apoE4–DPPC crystals at room temperature. Since apolipoproteins, including apoE, do not efficiently form complexes with DPPC spontaneously, we adapted the sodium cholate dialysis method for preparing complexes of apoA-I with DPPC (Sorci-Thomas et al., 1993 ▶). To maximize the chances of crystallization, our strategy was to make small homogeneous particles. First, this required optimization of the ratio of apoE to DPPC. The most homogeneous particles were at a sodium cholate concentration of 30 mg ml−1 and a 1:46 (E:DPPC) molar ratio (Fig. 1 ▶ a). Next, the sodium cholate:DPPC ratio was optimized. A molar ratio of 1.4:1 was optimal for the generation of homogeneous particles (Fig. 1 ▶ b). Cholate removal by high-temperature dialysis produces a varied range of lipoprotein particles; however, removal of excess cholate by incubation with Bio-beads at room temperature avoids the lipid phase transitions and results in more stable uniformly small particles. Since lipid-free apoE4 aggregates at high temperature and lipid-associated apoE does not, apoE4–DPPC particles were easily separated from unbound protein and uncomplexed DPPC by gel-filtration chromatography on Superdex 200.
Figure 1.
Nondenaturing gel analysis of optimized conditions for preparation of small homogeneous particles. (a) Varying the molar ratio of apoE4:DPPC. (b) Varying the molar ratio of sodium cholate:apoE4–DPPC.
As determined by negative-staining electron microscopy, the isolated particles measured 60 × 80 Å. Each particle contained two molecules of apoE4 as determined by chemical cross-linking with dimethyl suberimidate (data not shown), and has a phospholipid:protein ratio of 1:1(w:w). As demonstrated in a competitive LDL receptor-binding assay, the apoE4–DPPC particles were biologically active in binding to fibroblast LDL receptors, with a 50% displacement concentration of 0.6 µg ml−1 compared with the apoE4 DMPC standard of 0.07 µg ml−1. As expected, the binding activity of the apoE4–DPPC particles was lower than that of the DMPC particles owing to the presence of fewer apoE molecules per particle (two compared with four). LDL receptor-binding activity is influenced by the number of apoE molecules per phospholipid particle (Pitas et al., 1979 ▶).
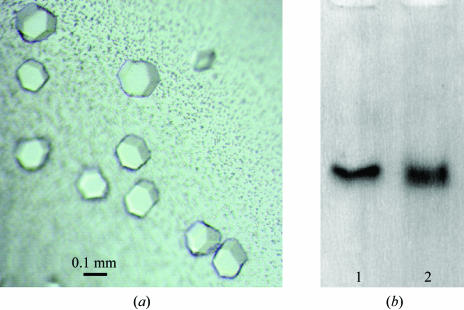
Based on follow-up from the initial Hampton screens, PEG 1500 and PEG 1000 were determined to be the precipitants of choice, as judged by reproducibility and crystal size. A pH grid identified the optimal pH as 5.8 with NaOAc and HEPES buffers. The additives 1,2,3-heptanetriol, 1,3-propanediol and cobalt chloride promoted crystal growth and size and decreased twinning, a common feature with these crystals. Crystals grown in PEG 1000 (24–25%), 20 mM sodium acetate pH 5.8 containing 1% 1,2,3-heptanetriol (crystal form A) are shown in Fig. 2 ▶(a). We proved that the crystals contained intact lipoprotein particles by dissolving a crystal and comparing its migration on a nondenaturing gel with that of a control apoE4–DPPC particle (Fig. 2 ▶ b).
Figure 2.
Crystallization of apoE4–DPPC. (a) Crystals were grown in PEG 1000, 20 mM sodium acetate pH 5.8, 33 mM 1,2,3-heptanetriol. (b) Immunoblot of dissolved apoE4–DPPC crystals. Lane 1, apoE4–DPPC control; lane 2, apoE4–DPPC crystals.
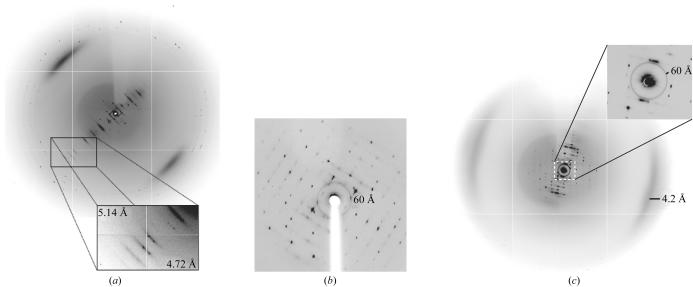
Three crystal forms of apoE4–DPPC have been analyzed in which sufficient data could be collected to determine the space group and unit cell (Table 1 ▶). For each of these crystal forms, the diffraction is anisotropic. For crystal form B, reflections at 4.5–7.5 Å can be observed in the first few frames when the crystals are oriented so that the 0h0 line is visible on the detector. However, the diffraction of these crystals decayed rapidly, so we are currently limited to collecting replicate reflections at 9 Å resolution. Crystal forms A and C diffract to 8–10 Å in this direction (Fig. 3 ▶ a). For all three forms, the highest resolution in the perpendicular direction is 8–12 Å (Fig. 3 ▶ b). For each of these crystals, we have collected data to 10–12 Å in order to minimize overloading of the low-resolution reflections. The space group and unit-cell parameters are summarized in Table 1 ▶.
Table 1. Diffraction data statistics for the three crystal forms of apoE4–DPPC.
| Crystal form | A | B | C |
|---|---|---|---|
| Unit-cell parameters | |||
| a (Å) | 79.0 | 62.3 | 79.7 |
| b (Å) | 111.4 | 79.3 | 113.6 |
| c (Å) | 99.4 | 169.3 | 186.0 |
| α (°) | 90.0 | 90.2 | 90.0 |
| β (°) | 113.0 | 90.1 | 90.0 |
| γ (°) | 90.0 | 110.5 | 90.0 |
| Completeness (%) | 98.5 | 23 | 99.6 |
| No. of reflections | 3061 | 2551 | 3061 |
| Unique reflections | 894 | 1446 | 894 |
| Rmerge | 3.9 (21) | 3.9 (24) | 3.9 (22) |
| Highest resolution shell (Å) | 11-10.0 | 9-8.5 | 11-10 |
| Maximum resolution† (Å) | 10.0 | 8.5 | 10.0 |
| Wavelength (Å) | 0.9797 | 1.078126 | 0.9797 |
| Space group | P21 | P1 | P2212, P21 |
| Matthews coefficient (Å3 Da−1) | 2.96 | 2.88 | 3.1 |
| Solvent content (%) | 58 | 57.3 | 60.3 |
| No. of molecules in ASU | 1 | 2 | 1 |
| NCS rotation (polar coordinates) (°) | (156.74, 0, 180) | None | None |
| Crystallization conditions | 24% PEG 1000, 20 mM NaOAc pH 5.8 | 24% PEG 1000, 20 mM NaOAc pH 5.8, 4 mM CoCl2 | 23% PEG 1500, 20 mM NaOAc pH 5.8, 4% 1,3-propanediol |
| Cryoprotectant | Ethylene glycol | Inositol | Ethylene glycol |
Maximum resolution is the resolution at which replicate reflections could be measured.
Figure 3.
Diffraction images of crystal form B. (a) 1° oscillation image showing the direction of maximal diffraction. The h00 axis is approximately perpendicular to the strong layer lines. (b) 1° oscillation image showing the direction of minimal diffraction. The image is cropped at 8 Å. (c) 0.25° oscillation image optimized to show the diffuse scattering. The ring at 60 Å and the diffuse arcs at 4.2 Å are most likely to result from the presence of DPPC in the crystals. Similar features are observed for unoriented DPPC bilayers (McIntosh & Simon, 1986 ▶; Small, 1986 ▶).
All crystal forms have strong diffuse scattering features at 60 and 4.2 Å (Fig. 3 ▶ c). These features are most likely to arise from the presence of lipid in the crystals, since similar features are observed for DPPC bilayers flash-frozen in the cryoprotectant conditions for crystal form B (data not shown). Earlier studies of stacked lamellar DPPC membranes at room temperature showed that the low-angle feature arises from the repeat distance between the phospholipid head groups, while the high-angle feature arises from packing between the glycerohydrocarbon tails (McIntosh & Simon, 1986 ▶).
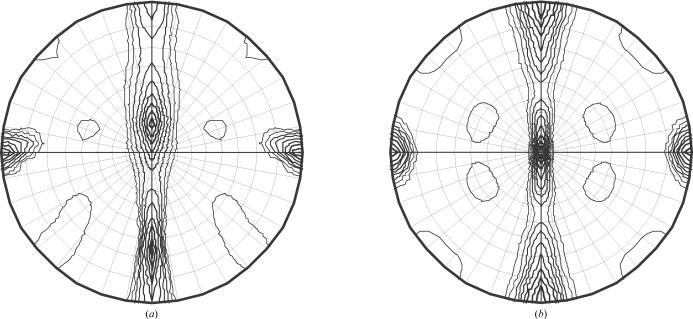
The symmetry of the two crystal forms with complete data sets (A and C) is consistent with the presence of two molecules of apoE per particle. The self-rotation function of crystal form B grown in the presence of 1,3-propanediol clearly shows 222 symmetry, indicating that the asymmetric unit contains one half of an apoE4–DPPC particle (Fig. 4 ▶ b). Crystal form A is clearly primitive monoclinic with systematic absences that suggest the presence of a 21 screw axis. It also has noncrystallographic twofold symmetry in addition to the crystallographic twofold as evidenced by the strong off-centre peak in the self-rotation function (Fig. 4 ▶ a). Since the asymmetric unit of crystal form A contains a complete apoE–DPPC particle, the apoE–DPPC particle must be twofold-symmetric to accommodate the noncrystallographic twofold. Therefore, both crystal forms suggest that the two apoE molecules adopt similar molecular envelopes on the surface of the apoE–DPPC particle in solution.
Figure 4.
Self-rotation functions. (a) χ = 180 ° section of the self-rotation function of crystal form A. (b) χ = 180° section of the self-rotation function of crystal form C.
Acknowledgments
We thank James Holten, Corie Ralston and Christine Trame for helpful discussions regarding data collection, Maureen Balestra for the LDL receptor-binding assays, John Carroll and Chris Goodfellow for graphics, Stephen Ordway and Gary Howard for editorial assistance, and Karina Fantillo for manuscript preparation. Diffraction analysis was performed at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division of the US Department of Energy under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. This work was supported in part by National Institutes of Health Grants AG20235 and HL64963 and the Gladstone Institutes.
References
- Arnold, K. S., Innerarity, T. L., Pitas, R. E. & Mahley, R. W. (1992). Lipoprotein Analysis. A Practical Approach, edited by C. A. Converse & E. R. Skinner, pp. 145–168. Oxford University Press.
- Corder, E. H., Saunders, A. M., Strittmatter, W. J., Schmechel, D. E., Gaskell, P. C., Small, G. W., Roses, A. D., Haines, J. L. & Pericak-Vance, M. A. (1993). Science, 261, 921–923. [DOI] [PubMed] [Google Scholar]
- Dong, L.-M., Innerarity, T. L., Arnold, K. S., Newhouse, Y. M. & Weisgraber, K. H. (1998). J. Lipid Res.39, 1173–1180. [PubMed] [Google Scholar]
- Drory, V. E., Birnbaum, M., Korczyn, A. D. & Chapman, J. (2001). J. Neurol. Sci.190, 17–20. [DOI] [PubMed] [Google Scholar]
- Fazekas, F., Strasser-Fuchs, S., Schmidt, H., Enzinger, C., Ropele, S., Lechner, A., Flooh, E., Schmidt, R. & Hartung, H.-P. (2000). J. Neurol. Neurosurg. Psychiatry, 69, 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innerarity, T. L., Pitas, R. E. & Mahley, R. W. (1979). J. Biol. Chem.254, 4186–4190. [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951). J. Biol. Chem.193, 265–275. [PubMed] [Google Scholar]
- McIntosh, T. J. & Simon, S. A. (1986). Biochemistry, 25, 4058–4066. [DOI] [PubMed] [Google Scholar]
- Mahley, R. W. (1988). Science, 240, 622–630. [DOI] [PubMed] [Google Scholar]
- Mahley, R. W., Ji, Z.-S., Brecht, W. J., Miranda, R. D. & He, D. (1994). Ann. NY Acad. Sci.737, 39–52. [DOI] [PubMed] [Google Scholar]
- Mayeux, R., Ottman, R., Maestre, G., Ngai, C., Tang, M.-X., Ginsberg, H., Chun, M., Tycko, B. & Shelanski, M. (1995). Neurology, 45, 555–557. [DOI] [PubMed] [Google Scholar]
- Pitas, R. E., Innerarity, T. L., Arnold, K. S. & Mahley, R. W. (1979). Proc. Natl Acad. Sci. USA, 76, 2311–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses, A. D. & Saunders, A. M. (1997). Ann. NY Acad. Sci.826, 200–212. [DOI] [PubMed] [Google Scholar]
- Saunders, A. M., Strittmatter, W. J., Schmechel, D., St George-Hyslop, P. H., Pericak-Vance, M. A., Joo, S. H., Rosi, B. L., Gusella, J. F., Crapper-MacLachlan, D. R., Alberts, M. J., Hulette, C., Crain, B., Goldgaber, D. & Roses, A. D. (1993). Neurology, 43, 1467–1472. [DOI] [PubMed] [Google Scholar]
- Slooter, A. J. C., Tang, M.-X., van Duijn, C. M., Stern, Y., Ott, A., Bell, K., Breteler, M. M. B., Van Broeckhoven, C., Tatemichi, T. K., Tycko, B., Hofman, A. & Mayeux, R. (1997). J. Am. Med. Assoc.277, 818–821. [DOI] [PubMed]
- Small, D. M. (1986). The Physical Chemistry of Lipids. From Alkanes to Phospholipids, pp. 512–517. New York: Plenum Press.
- Sorci-Thomas, M., Kearns, M. W. & Lee, J. P. (1993). J. Biol. Chem.268, 21403–21409. [PubMed] [Google Scholar]
- Strittmatter, W. J., Saunders, A. M., Schmechel, D., Pericak-Vance, M., Enghild, J., Salvesen, G. S. & Roses, A. D. (1993). Proc. Natl Acad. Sci. USA, 90, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, G. M., Nicoll, J. A. R., Murray, G. & Fiddes, M. (1997). Lancet, 350, 1069–1071. [DOI] [PubMed] [Google Scholar]
- Weisgraber, K. H. (1994). Adv. Protein Chem.45, 249–302. [DOI] [PubMed] [Google Scholar]