The mammalian Pax6 paired domain has been cocrystallizaed with a 25 bp DNA fragment of the Pax6 gene enhancer.
Keywords: Pax6 paired domain, Pax6 gene enhancer, Pax-family proteins
Abstract
Pax6 is a member of the Pax family of transcription factors and is essential for eye development. Pax6 has two DNA-binding domains: the paired domain and the homeodomain. The Pax6 paired domain is involved in Pax6 gene autoregulation by binding to its enhancer. In this study, crystallization and preliminary X-ray diffraction analysis of the mammalian Pax6 paired domain in complex with the Pax6 gene enhancer was attempted. The Pax6 paired domain complexed with an optimized 25 bp DNA fragment was crystallized by the hanging-drop vapour-diffusion method. The crystal diffracted synchrotron radiation to 3.0/3.7 Å resolution and belongs to the monoclinic space group P21, with unit-cell parameters a = 62.21, b = 70.69, c = 176.03 Å, β = 90.54°. Diffraction data were collected to 3.7 Å resolution.
1. Introduction
The Pax-family proteins are defined by the presence of a highly conserved paired box which corresponds to a 128-amino-acid paired domain (PD). They are transcription factors involved in developmental processes such as the formation of the central nervous system (Dahl et al., 1997 ▶). In the mammalian genome, nine Pax genes have been identified and divided into four subgroups based on their sequence homology and structural similarity: Pax1/9, Pax2/5/8, Pax3/7 and Pax4/6 (Eccles et al., 2002 ▶).
Pax6 is directly related to eye and central nervous system development in both vertebrates and invertebrates (Halder et al., 1995 ▶; Treisman, 2004 ▶; Simpson & Price, 2002 ▶). Mutations in the Pax6 gene cause aniridia in humans (Jordan et al., 1992 ▶), small eye in mice (Hill et al., 1991 ▶) and eyelessness in Drosophila (Quiring et al., 1994 ▶). In Drosophila, ectopic expression of either the Drosophila or mouse Pax6 gene can induce the formation of ectopic eyes (Halder et al., 1995 ▶). These findings suggest that Pax6 is the key control gene in eye development. The two DNA-binding domains, the PD and the homeodomain (HD), and the C-terminal transactivation domain, which is rich in proline, serine and threonine, are involved in the Pax6-mediated transcriptional activation of target genes. Several Pax6 target gene candidates have been identified, including crystallins (Cvekl, Kashanchi et al., 1995 ▶; Cvekl, Sax et al., 1995 ▶; Richardson et al., 1995 ▶; Duncan et al., 1998 ▶), rhodopsin (Sheng et al., 1997 ▶), c-maf (Sakai et al., 2001 ▶), neurogenin-2 (Scardigli et al., 2001 ▶) and neural cell-adhesion molecule L1 (Meech et al., 1999 ▶). In the chicken δ1-crystallin enhancer, Pax6 functions together with Sox2, another transcription factor involved in developmental control (Kamachi et al., 2001 ▶). The PD and the HD have both independent and cooperative DNA-binding abilities (Jun & Desplan, 1996 ▶). Consensus DNA sequences bound to each domain have been proposed based on in vitro binding experiments designed to select recognition sites for the PD and the HD (Epstein et al., 1994 ▶; Wilson et al., 1996 ▶). The helix–turn–helix motif, a well known DNA-binding element, is present in both the PD and the HD (Xu et al., 1999 ▶; Wilson et al., 1995 ▶). Indeed, the PD structure has two helix–turn–helix motifs connected by a linker. These motifs bind to the major groove of an optimal Pax6 PD-binding DNA sequence in a symmetric manner (Xu et al., 1999 ▶).
Recently, a novel Pax6 protein-responsive DNA element termed LE9 (52 bp in length) was identified (Aota et al., 2003 ▶). With 100% identity between human and mouse, LE9 is an enhancer sequence of the Pax6 gene itself and resides in its head surface ectoderm-specific enhancer region. The Pax6 PD binds to the distal half site of LE9 DNA, while the proximal half site of LE9 can bind the Sox2 HMG domain. This evidence suggested that Pax6 and LE9 directly interact in the autoregulation of the Pax6 gene. In addition, the Pax6-responsive enhancer activity of a single LE9 DNA was stronger than that of the optimal Pax6 PD-binding DNA. Although the LE9 DNA sequence is comparable with the optimal Pax6 PD-binding DNA sequence, there are some significant differences between them. The DNA-sequence identity in the Pax6-binding region (approximately 20 base pairs; Xu et al., 1999 ▶) was at most 60%, even when we allowed one base-pair deletion in the LE9 sequence (Aota et al., 2003 ▶). Thus, the precise interaction mode of the Pax6 PD with LE9 DNA should differ from that observed in the crystal structure of the Pax6 PD in complex with the optimal DNA: (i) Pax6 PD may bind to LE9 DNA with a very similar protein conformation, but recognize a different DNA sequence, or (ii) Pax6 PD may bind to the LE9 sequence with a different protein conformation in order to compensate for the one base-pair deletion of the LE9 sequence.
To understand the functional structure of the Pax6 PD in DNA recognition, we have crystallized the PD of the mammalian Pax6 protein in complex with a 25 bp duplex derived from the PD-binding site of LE9 which was optimized for crystallization.
2. Materials and methods
2.1. Protein expression and purification
To express the Pax6 PD as a GST-fusion protein, the human cDNA fragment encoding residues 4–136 of Pax6 was cloned into the pGEX-6p-2 vector (Amersham Biosciences). The entire amino-acid sequences of both human and mouse Pax6, including the paired domain, are 100% identical. The constructed plasmid vector was transformed into Escherichia coli strain BL21 (DE3) CodonPlus RIL cells (Stratagene) for the expression of a GST-fusion protein. The transformed cells were grown in 1 l LB medium containing 100 µg ml−1 ampicillin at 310 K. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM when the OD600 reached 0.3 at 310 K. After further cultivation at 293 K for 18 h, the cells were harvested and resuspended in buffer A (40 mM HEPES pH 7.5, 5 mM DTT, 1 mM EDTA, 200 mM NaCl) with a protease-inhibitor cocktail (Roche) and then lysed by sonication. The lysate was clarified by centrifugation at 36 000g for 30 min (277 K). Polyethyleneimine (0.15% final concentration) was added to the supernatant and after incubation for 10 min at 277 K the mixture was centrifuged at 36 000g for 20 min (277 K). After the proteins had been precipitated by ammonium sulfate (80% saturation), the pellet was collected by centrifugation and dissolved in buffer A. The solution was loaded onto a GSTrap column (Amersham Biosciences) and the GST-Pax6PD proteins were eluted with 10 mM reduced glutathione. To remove the N-terminal GST tag, Precision protease (10 U per milligram of protein, Amersham Biosciences) was added to the protein solution and the sample was incubated for 16 h at 277 K. The GST tag was separated from Pax6 PD using cation-exchange chromatography. The incubated sample was loaded onto a HiTrap-SP column (Amersham Biosciences) and the column was developed using a linear gradient of 0.2–1 M NaCl in buffer A. Pax6 PD was purified to homogeneity with a yield of approximately 2.5 mg per litre of bacterial cells. The purified protein was concentrated for crystallization to 15 mg ml−1 using an AmiconUltra filter (MWCO 5000, Amicon) in 5 mM NaOAc pH 5.0.
2.2. Crystallization
The DNA oligonucleotides (Fig. 1 ▶ a) were purchased from Bex (Tokyo, Japan). They were designed to form conformationally continuous DNA helices through a 5′ two base pair overlap in the crystal (each end was AA or TT). After the single-stranded DNAs had been heated at 371 K for 10 min in 5 mM NaOAc pH 5.0, they were annealed by gradual cooling to 277 K. The protein–DNA complexes were formed by mixing equimolar amounts of the protein and the DNA. Crystallization was carried out by the hanging-drop vapour-diffusion method at 277 K. Crystals were obtained when a 1.6 µl aliquot of each protein–DNA complex was mixed with 0.8 µl of a reservoir solution containing 20% PEG 8000 and 200 mM MgOAc pH 6.5 (Hampton Research Crystal Screen kit No. 18). The crystals appeared in a few days and were allowed to grow for several days.
Figure 1.
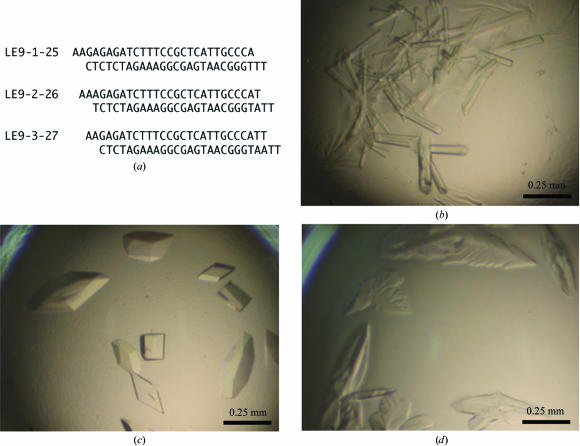
(a) DNA sequences used in cocrystallization with the Pax6 paired domain. (b) Crystals of the Pax6 paired domain in complex with LE9-1-25 DNA. The precipitant buffer contained 18% PEG 8000 and 200 mM MgOAc pH 6.5. (c) Crystals with LE9-2-26 DNA that diffracted X-rays, as shown in Fig. 2 ▶. (d) Crystals with LE9-3-27 DNA.
2.3. X-ray data collection and processing
For X-ray diffraction measurements at 100 K, the crystals were cryoprotected by soaking into the aforementioned crystallization buffer supplemented with 20%(v/v) glycerol and were flash-frozen in a cold nitrogen-gas stream. Diffraction data were collected at beamline BL38B1 of SPring-8 (Hyogo, Japan), which is equipped with a Rigaku Jupiter 210 CCD detector. The oscillation angle per image was 1.0°, the crystal-to-detector distance was 260 mm and the wavelength was 1.0000 Å. A total of 180 frames were recorded, with 15 s of exposure per image. The collected data set was indexed and scaled with the HKL2000 program (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
We were able to obtain crystals of the Pax6 PD in complex with the Pax6 gene enhancer using PEG 8000 as a precipitant at 277 K. Interestingly, the DNA sequence greatly affected the shape and quality of the crystals with the Pax6 PD. Diamond-shaped crystals of the best quality were obtained with the DNA 2–26 sequence (25 bp duplex) of LE9 (LE9-2-26; Fig. 1 ▶ c; Aota et al., 2003 ▶). When the DNA LE9-1-25 sequence, a version shifted by only one base pair, was crystallized with the Pax6 PD, the crystals were rod-shaped and did not diffract X-rays (Fig. 1 ▶ b). Moreover, when the DNA LE9-3-27 sequence was used, the shapes of the crystals were irregular and they diffracted X-rays poorly (Fig. 1 ▶ d). In an electrophoretic mobility shift assay, all three DNAs bound to the Pax6 PD with similar binding affinity (data not shown). These results suggest that the DNA sequence is crucial for the regular packing of the protein and DNA molecules within the crystal lattice.
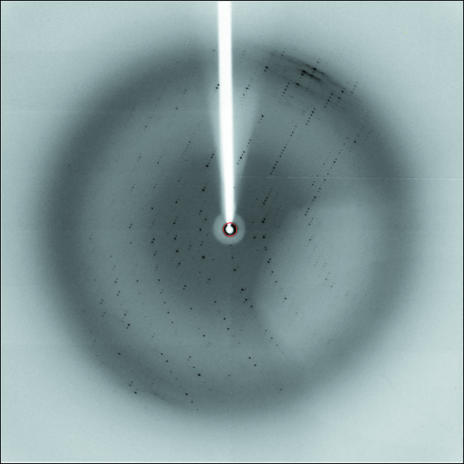
X-ray diffraction experiments for the diffracting crystals were performed using synchrotron radiation at SPring-8 (Fig. 2 ▶). Although the crystals tended to diffract X-rays poorly and anisotropically, the best crystal diffracted to 3.0/3.7 Å resolution. The crystals belong to the monoclinic space group P21, with unit-cell parameters a = 62.21, b = 70.69, c = 176.03 Å, β = 90.54°. The diffraction data set was collected isotropically to 3.7 Å resolution. The data-collection statistics are summarized in Table 1 ▶. Considering the DNA length, the DNA axis should be almost parallel to the crystallographic c axis and a self-rotation function supported this interpretation (data not shown). Assuming the presence of four complex molecules per asymmetric unit, the Matthews coefficient (V M) was estimated to be 3.07 Å3 Da−1, corresponding to a solvent content of approximately 58%. On the other hand, the fragile nature and anisotropic diffraction implies a larger V M and a higher solvent content.
Figure 2.
X-ray diffraction pattern of the best crystal (the Pax6 paired domain complexed with LE9-2-26 DNA). The diffraction data were recorded at 100 K on a Rigaku Jupiter 210 CCD at SPring-8, with a camera distance of 260 mm, an oscillation angle of 1.0°, a wavelength of 1.0000 Å and an exposure time of 15 s. The frame edge is at 2.6 Å resolution.
Table 1. X-ray data-collection statistics.
Values in parentheses refer to the highest resolution shell.
| Wavelength (Å) | 1.0000 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 62.21, b = 70.69, c = 176.03, β = 90.54 |
| Resolution range (Å) | 50.0–3.7 (3.83–3.70) |
| Observed reflections | 55646 (3993) |
| Unique reflections | 15550 (1260) |
| Redundancy | 3.6 (3.2) |
| Completeness (%) | 94.8 (77.6) |
| Rmerge† (%) | 7.1 (16.2) |
| I/σ(I) | 9.9 (3.0) |
R
merge = 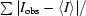
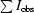 , where I
obs is the intensity measurement for a given reflection and 〈I〉 is the average intensity for multiple measurements of this reflection.
, where I
obs is the intensity measurement for a given reflection and 〈I〉 is the average intensity for multiple measurements of this reflection.
Structure determination by molecular-replacement techniques, using the equivalent domain of the Pax6 PD and an optimal DNA sequence (PDB code 6pax; Xu et al., 1999 ▶) as a probe, were unsuccessful. Therefore, we are currently performing structure determination by the isomorphous replacement method using heavy-atom derivatives and also improving the quality of the crystals in order to determine the functional interaction mode between the Pax6 PD and DNA at an atomic level.
Acknowledgments
We thank Ms Tomoko Ishigaki for assistance with sample preparation and Drs Kazuya Hasegawa and Hisanobu Sakai for use of the facilities at SPring-8. This work was supported by grants from New Energy and Industrial Technology Development Organization (NEDO, Japan).
References
- Aota, S., Nakajima, N., Sakamoto, R., Watanabe, S., Ibaraki, N. & Okazaki, K. (2003). Dev. Biol.257, 1–13. [DOI] [PubMed] [Google Scholar]
- Cvekl, A., Kashanchi, F., Sax, C. M., Brady, J. N. & Piatigorsky, J. (1995). Mol. Cell. Biol.15, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl, A., Sax, C. M., Li, X., McDermott, J. B. & Piatigorsky, J. (1995). Proc. Natl Acad. Sci. USA, 92, 4681–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, E., Koseki, H. & Balling, R. (1997). BioEssays, 19, 755–765. [DOI] [PubMed] [Google Scholar]
- Duncan, M. K., Haynes, J. I. II, Cvekl, A. & Piatigorsky, J. (1998). Mol. Cell. Biol.18, 5579–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles, M. R., He, S., Legge, M., Kumar, R., Fox, J., Zhou, C., French, M. & Tsai, R. W. (2002). Int. J. Dev. Biol.46, 535–544. [PubMed] [Google Scholar]
- Epstein, J., Cai, J., Glaser, T., Jepeal, L. & Maas, R. (1994). J. Biol. Chem.269, 8355–8361. [PubMed] [Google Scholar]
- Halder, G., Callaerts, P. & Gehring, W. J. (1995). Science, 267, 1788–1792. [DOI] [PubMed] [Google Scholar]
- Hill, R. E., Favor, J., Hogan, B. L., Ton, C. C., Saunders, G. F., Hanson, I. M., Prosser, J., Jordan, T., Hastie, N. D. & van Heyningen, V. (1991). Nature (London), 354, 522–525. [DOI] [PubMed] [Google Scholar]
- Jordan, T., Hanson, I., Zaletayev, D., Hodgson, S., Prosser, J., Seawright, A., Hastie, N. & van Heyningen, V. (1992). Nature Genet.1, 328–332. [DOI] [PubMed] [Google Scholar]
- Jun, S. & Desplan, C. (1996). Development, 122, 2639–2650. [DOI] [PubMed] [Google Scholar]
- Kamachi, Y., Uchikawa, M., Tanouchi, A., Sekido, R. & Kondoh, H. (2001). Genes Dev.15, 1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech, R., Kallunki, P., Edelman, G. M. & Jones, F. S. (1999). Proc. Natl Acad. Sci. USA, 96, 2420–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Quiring, R., Walldorf, U., Kloter, U. & Gehring, W. J. (1994). Science, 265, 785–789. [DOI] [PubMed] [Google Scholar]
- Richardson, J., Cvekl, A. & Wistow, G. (1995). Proc. Natl Acad. Sci. USA, 92, 4676–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, M., Serria, M. S., Ikeda, H., Yoshida, K., Imaki, J. & Nishi, S. (2001). Nucleic Acids Res.29, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli, R., Schuurmans, C., Gradwohl, G. & Guillemot, F. (2001). Neuron, 31, 203–217. [DOI] [PubMed] [Google Scholar]
- Sheng, G., Thouvenot, E., Schmucker, D., Wilson, D. S. & Desplan, C. (1997). Genes Dev.11, 1122–1131. [DOI] [PubMed] [Google Scholar]
- Simpson, T. I. & Price, D. J. (2002). BioEssays, 24, 1041–1051. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E. (2004). Development, 131, 3823–3827. [DOI] [PubMed] [Google Scholar]
- Wilson, D. S., Guenther, B., Desplan, C. & Kuriyan, J. (1995). Cell, 82, 709–719. [DOI] [PubMed] [Google Scholar]
- Wilson, D. S., Sheng, G., Jun, S. & Desplan, C. (1996). Proc. Natl Acad. Sci. USA, 93, 6886–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. E., Rould, M. A., Xu, W., Epstein, J. A., Maas, R. L. & Pabo, C. O. (1999). Genes Dev.13, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]