The carbohydrate-binding component (VP8*64–223) of the human Wa rotavirus spike protein has been overexpressed in E. coli, purified and crystallized in two different crystal forms. X-ray diffraction data have been collected that have enabled determination of the Wa VP8*64–223 structure by molecular replacement.
Keywords: human Wa rotavirus, carbohydrate-binding protein, VP8*
Abstract
Rotaviruses exhibit host-specificity and the first crystallographic information on a rotavirus strain that infects humans is reported here. Recognition and attachment to host cells, leading to invasion and infection, is critically linked to the function of the outer capsid spike protein of the rotavirus particle. In some strains the VP8* component of the spike protein is implicated in recognition and binding of sialic-acid-containing cell-surface carbohydrates, thereby enabling infection by the virus. The cloning, expression, purification, crystallization and initial X-ray diffraction analysis of the VP8* core from human Wa rotavirus is reported. Two crystal forms (trigonal P3221 and monoclinic P21) have been obtained and X-ray diffraction data have been collected, enabling determination of the VP8*64–223 structure by molecular replacement.
1. Introduction
Host-cell attachment and entry by rotaviruses involves several possible receptors, but their exact roles and the overall mechanism of this multi-step process remain unresolved. Rotaviruses require delivery of their complete icosahedral particle across a membrane and into the cytoplasm, where calcium-dependent uncoating of the virion occurs to give the transcriptionally active double-layered particle and initiation of viral gene expression. The virion comprises a triple-layered protein structure surrounding a genome of 11 segments of double-stranded RNA (Estes, 2001 ▶). The outermost layer comprises two structural proteins, VP4 and VP7; both elicit neutralizing antibodies and independently define serotype specificity and are also involved in initial host-cell interactions crucial to rotavirus infectivity (Coulson et al., 1997 ▶; Estes & Cohen, 1989 ▶; Estes, 2001 ▶; Prasad & Chiu, 1994 ▶). The viral haemagglutinin VP4 forms a spike that exists in a dimeric or possibly a trimeric state and extends up to 100 Å from the virion surface. Infectivity is increased by proteolytic cleavage of VP4, producing an N-terminal carbohydrate-binding domain (VP8*) and a C-terminal fragment (VP5*) that both remain associated with the virion. It has been postulated that many, if not all, rotaviruses initially interact with a sialic acid-containing cellular glycoconjugate prior to recognition of additional cell-surface receptors, including integrins (Jolly et al., 2000 ▶; Hewish et al., 2000 ▶). The infectious triple-layered particle binds gangliosides, unlike the double-layered particle (Delorme et al., 2001 ▶), demonstrating that one or both of VP4 and VP7 are involved in oligosaccharide binding.
Human and animal rotavirus strains have been categorized according to their ability to bind and infect cultured cells that have been treated with sialidases, which remove sialic acids such as N-acetylneuraminic acid from cell-surface glycoconjugates (Ciarlet & Estes, 1999 ▶; Fukudome et al., 1989 ▶). Rotaviruses that show this dependence are termed sialidase-sensitive. Sialidase sensitivity correlates with the VP4 serotype (Ciarlet et al., 2002 ▶). In experiments using sialidases that only cleave terminal (but not internal) sialic acid residues, an indication is given as to whether infection is dependent upon recognition and binding of a terminal sialic acid residue on the glycoconjugate. Interestingly, rotaviruses isolated from humans are mostly sialidase-insensitive and therefore either do not recognize sialic acids at all or recognize internal sialic acids within the oligosaccharide chains. Currently, the question of whether there is an absolute requirement for sialic acid recognition to enable infection is unresolved. However, there is increasing evidence to support human rotaviruses having the capacity to recognize sialic acids located internally within oligosaccharide chains (Delorme et al., 2001 ▶; Guerrero et al., 2000 ▶; Guo et al., 1999 ▶; Jolly et al., 2000 ▶). This is exemplified by the involvement of the sialic acid-containing ganglioside GM1 on the host-cell surface in human rotavirus infection (Guo et al., 1999 ▶). Mechanisms controlling the remarkable host-specificity demonstrated by rotaviruses are not known, although some commonality in the multiple cell-surface interactions leading to virions binding to cells does exist between human and lower animal strains (Guo et al., 1999 ▶). However, there also are substantial differences in the structural components of the actual glycoconjugates recognized. Investigation of the determinants of host-specificity is of importance in the design of rotavirus-specific therapeutics.
Rotaviruses are the most important viral-induced cause of severe gastroenteritis and resulting diarrhoea in the young of most mammalian species, via infection of epithelial cells in the small intestine (Estes, 2001 ▶). An estimated average of 440 000 deaths annually of children less than 5 y old (Parashar et al., 2003 ▶) occurs. The rotavirus most often detected in ill children worldwide is of VP4 serotype 1A and VP7 serotype 1 (Santos & Hoshino, 2005 ▶). Human rotavirus strain Wa is the prototype for this serotype. Given the impact of rotavirus disease and the proven importance of VP4 in recognition of host cells during infection, the VP8* subunit of VP4 is an important drug-design target. To date, the only structural information on rotavirus VP8* carbohydrate-binding domains relates to the sialidase-sensitive strains of monkey (RRV; Dormitzer et al., 2002 ▶) and our investigation into porcine CRW-8 (Scott et al., 2005 ▶). To provide new insight into carbohydrate recognition by rotaviruses and their host-specificity, we report here the cloning, overexpression, purification and determination of the X-ray crystallographic structure of VP8*64–223 from the sialidase-insensitive human rotavirus strain Wa.
2. Experimental procedures and results
2.1. Cloning, expression and purification of human Wa VP8*64–223
2.1.1. Cloning
Viral dsRNA was extracted from rotavirus strain Wa (passage 10) as previously described (Dyall-Smith & Holmes, 1984 ▶). This dsRNA was used as a template for cDNA synthesis using the Superscript First-Strand Synthesis System (Invitrogen) for reverse transcription–polymerase chain reaction (RT–PCR). The primer used for RT was 5′-GTAGAAACCCTCCATTATAGC-3′ and the products of the RT reaction were then used as templates for two nested PCR rounds using the Expand High Fidelity PCR system (Roche) to amplify the VP8* gene fragment for cloning into the bacterial expression vector pGEX-4T-1 (Amersham Biosciences). In the first round the forward primer 5′-GGCTATAAAATGGCTTCACTC-3′ was used along with the same primer used in the RT reaction as the reverse primer. The product from this reaction was then used as a template in a second round of PCR using the forward primer 5′-CGCGGATCCATGTTAGATGGTCCTTATCAGCC-3′ and the reverse primer 5′-GGAATTCTCACAGACCATTATTAATATATTC-3′. These primers contained BamHI and EcoRI restriction sites (bold) to enable insertion of the PCR product into pGEX-4T-1. Following digestion, PCR products were ligated into pGEX-4T-1, yielding pGEX-Wa-VP8*, which encoded amino-acid residues 64–223 of Wa VP4 fused to the C-terminus of GST. The integrity of the inserted VP8* gene fragment was assessed by DNA sequencing. The predicted amino-acid sequence of pGEX-Wa-VP8* was identical to the published Wa VP4 sequence except for leucine substituting a phenylalanine as position 163 (accession No. L34161, Entrez Database, NCBI, NIH).
2.1.2. Expression and purification
Escherichia coli strain BL21 DE3 transformed with the pGEX-Wa-VP8* plasmid was grown at 310 K in Luria–Bertani (LB) media supplemented with 150 µg ml−1 ampicillin. Upon reaching an OD600 of 0.6, the cultures were incubated at 298 K for 1 h and then induced with 1 mM isopropyl α-d-thiogalactopyranoside (BioVectra DCL). Cells were harvested 4 h after induction by centrifugation at 8000g for 10 min and frozen. Frozen cell pellets were thawed in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 pH 7.4 (PBS) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Roche Diagnostics). The cells were lysed with 3 mg ml−1 lysozyme (Fluka) supplemented with 1%(w/v) Triton X-100 and 20 µg ml−1 DNaseI (Roche Diagnostics). Cell debris was removed by centrifugation at 20 000g for 30 min and the supernatant was passed over a glutathione-Sepharose column (Amersham Pharmacia Biotech). The column was then washed with 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM CaCl2 (TNC) and bound GST-fusion protein was digested with 10 µg ml−1 TPCK-treated trypsin (Worthington Biochemical) for 2 h at room temperature. A benzamidine-Sepharose (Amersham-Pharmacia Biotech) column pre-equilibrated with TNC was connected in series with the glutathione-Sepharose column. The cleaved protein was eluted with 20 mM NaPO4 pH 7.5, 1 M NaCl and 1 mM PMSF and 2.5 mM benzamidine was added to the eluant. The protein was dialysed against 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA (TNE). This protein was concentrated to ∼2 ml using an Amicon Ultrapure-15 centrifugal filter device (Millipore) and subjected to size-exclusion chromatography over Sephacryl S100 resin (Sigma) equilibrated in TNE. Purified protein was analysed for homogeneity by SDS–PAGE (Fig. 1 ▶) and dynamic light scattering (DLS) using a CoolBatch Plus 90T instrument (Precision Detectors). DLS indicated a monomodal distribution of the purified Wa VP8*64–223 protein, with an observed hydrodynamic radius (R h) of 1.46 nm (the calculated R h is 1.47 nm) and 18.7% polydispersity, which falls within the range (15–30%) indicative of good homogeneity.
Figure 1.
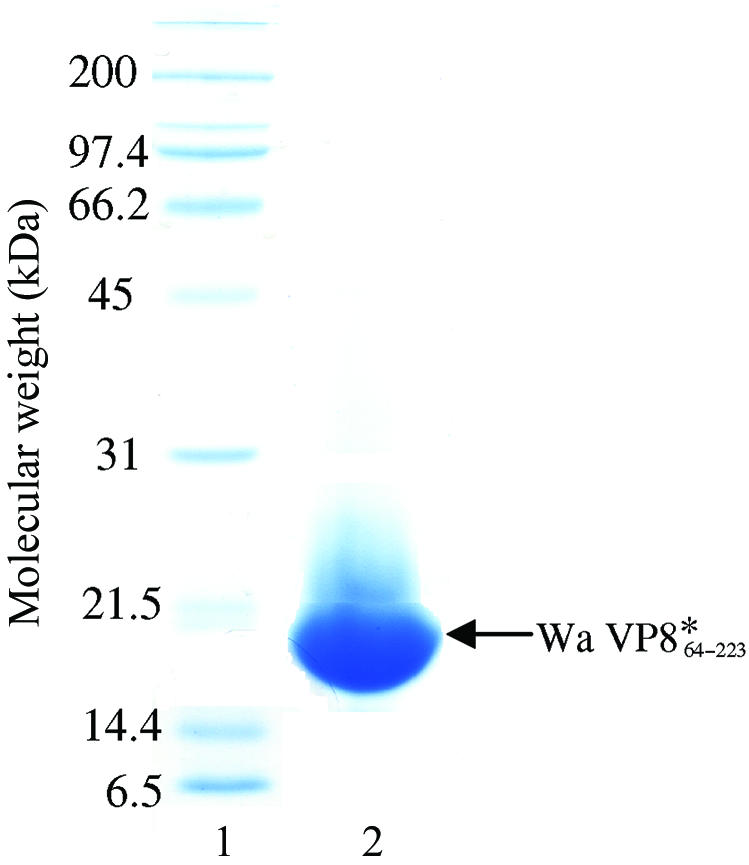
SDS–PAGE (12%, stained with Coomassie blue) analysis of human Wa VP8*64–223 (calculated molecular weight of 18 380 Da). Lane 1 is the ladder. Lane 2 is the protein sample; 105 µg was loaded (the protein sample was at a concentration of 105.5 mg ml−1).
2.2. Crystallization
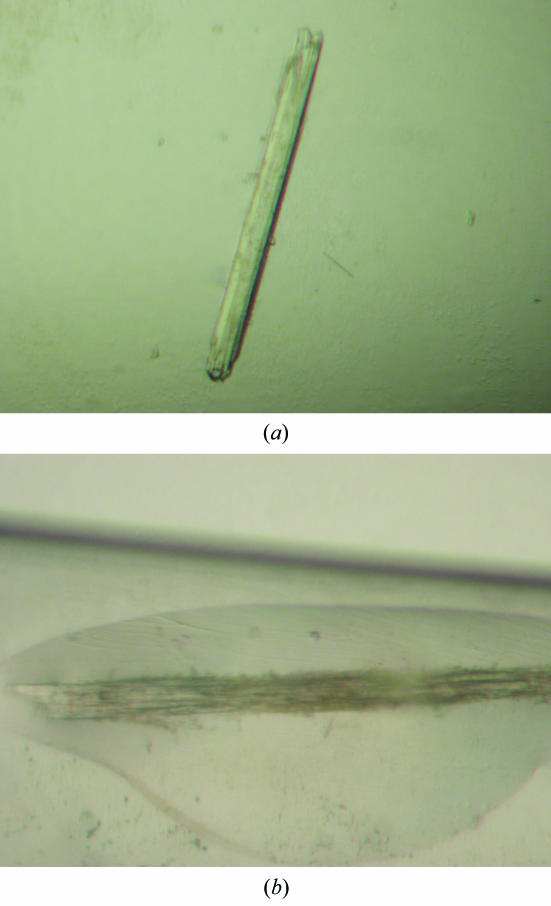
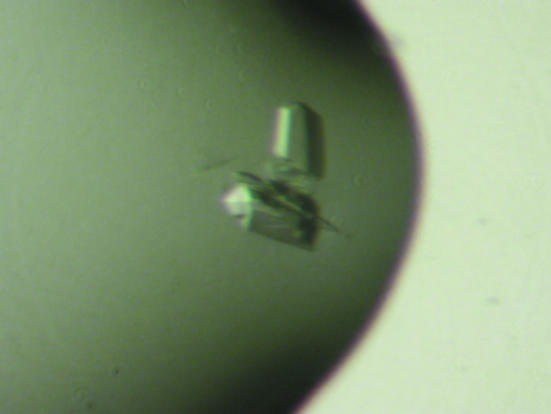
Crystallization experiments in the presence and absence of the sialic acid derivative methyl α-d-N-acetylneuraminide [which bound to the RRV (Dormitzer et al., 2002 ▶) and porcine CRW-8 VP8* structures (Scott et al., 2005 ▶)] were performed using a sparse-matrix approach (Jancarik & Kim, 1991 ▶) with Crystal Screen kits (Hampton Research) at 293 K and the hanging-drop vapour-diffusion technique. Crystals formed under two conditions, one of which (form A) produced crystals in the presence and absence of ligand. Wa VP8*64–223 (30 mg ml−1 in TNE) was premixed with methyl α-d-N-acetylneuraminide at 92 mM concentration (protein:ligand ratio of 1:50). Drops consisted of equal volumes of this mixture and reservoir solution comprising 25%(w/v) PEG 4000, 20%(v/v) 2-propanol, 0.1 M sodium citrate pH 5.6. Triangular rod-shaped crystals (form A) grew after 5 d to typical dimensions of 0.25 × 0.1 × 0.08 mm (Fig. 2 ▶). In the absence of ligand, crystals grew after optimization of the drop contents whilst retaining identical reservoir solution. Specifically, the drops comprised 1.6 µl protein solution (20 mg ml−1 in TNE) and 1.3 µl of a solution containing 14.4%(w/v) PEG 4000, 0.1 M sodium citrate pH 5.6, 20% 2-propanol and 0.3 µl of the additive ethylene glycol. Sequential macroseeding over ten weeks generated a crystal suitable for X-ray diffraction data collection (Fig. 3 ▶ a). The crystals grew fine surface rods as exhibited by the crystal shown in Fig. 3 ▶(b) at the time of data collection. Crystals of form B formed under alternative conditions consisting of the combination of an equal volume of premixed protein–ligand solution (protein at 17 mg ml−1 and methyl α-d-N-acetylneuraminide at 53 mM concentration; a protein:ligand ratio of 1:50) and reservoir solution comprising 30%(w/v) PEG 4000, 0.2 M ammonium acetate, 0.1 M sodium citrate pH 5.6. Crystals appeared after three months and grew to dimensions of 0.25 × 0.15 × 0.1 mm (Fig. 4 ▶).
Figure 2.
Crystals (form A) of human Wa VP8*64–223 grown in the presence of ligand via the hanging-drop vapour-diffusion method. Typical crystal size is 0.25 × 0.1 × 0.08 mm.
Figure 3.
Crystal (form A) of human Wa VP8*64–223 grown in the absence of ligand. The crystal for which the room-temperature X-ray diffraction data set was collected is shown (a) two weeks before data collection and (b) at the time of data collection (mounted in a quartz capillary), exhibiting surface crystals. The crystal is of dimensions 0.65 × 0.05 × 0.05 mm.
Figure 4.
Crystals (form B) of human Wa VP8*64–223 grown in the presence of ligand via the hanging-drop vapour-diffusion method. Crystal dimensions are ∼0.25 × 0.15 × 0.1 mm.
2.3. X-ray diffraction analysis and structure determination.
X-ray diffraction data were collected on the FIP-BM30A station of the European Synchrotron Radiation Facility (ESRF; 100 K, λ = 0.9794 Å) using a MAR CCD detector for both crystal forms of Wa VP8*64–223 grown in the presence of ligand. A room-temperature (295 K) data set from a form A crystal grown without ligand was collected on a ProteumR (Bruker AXS, Madison, WI, USA) diffractometer with a MacScience M06XCE rotating-anode generator equipped with a SMART6000 CCD detector. Indexing and processing of the synchrotron data sets were performed with MOSFLM (Leslie, 1992 ▶) and SCALA (CCP4 suite; Collaborative Computational Project, Number 4, 1994 ▶). Rotating-anode data were integrated using SAINT (v.1.0: Bruker AXS Inc., Madison, WI, USA) and scaled with PROSCALE (v.7.10: Bruker AXS Inc., Madison, WI, USA). For molecular-replacement purposes, a homology model of human Wa VP8*64–223 was built using MODELLER (Sali & Blundell, 1993 ▶) based upon amino-acid sequence alignment with rhesus rotavirus (RRV) VP8* (45% amino-acid sequence identity) and the three-dimensional protein structure of RRV VP8*62–224 (Dormitzer et al., 2002 ▶). Molecular replacement was performed using AMoRe (Navaza, 1994 ▶). Rotation- and translation-function calculations utilized data within the range 8–3 Å and structure refinement was performed using REFMAC (Murshudov et al., 1997 ▶).
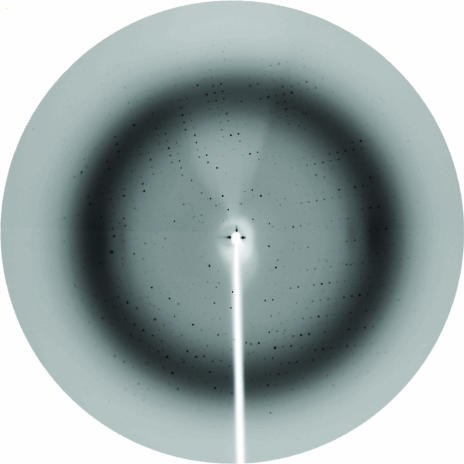
X-ray diffraction data to 2.5 Å (Fig. 5 ▶, Table 1 ▶) were collected from a triangular rod-shaped crystal (form A; Fig. 2 ▶) after being dipped into a cryoprotectant solution containing 5%(v/v) glycerol and reservoir solution and flash-frozen in liquid nitrogen. Assuming the presence of one molecule per asymmetric unit of the trigonal system (space group P3221), the Matthews coefficient (Matthews, 1968 ▶) V M is 3.1 Å3 Da−1 and the solvent content is 60%. Molecular replacement gave the highest rotation-function peak with an R factor of 54.9% and a correlation coefficient of 20.2%, marginally distinct from the next highest peak (R factor 55.4%, correlation coefficient 18.5%). Translation-function solutions for search models rotated to the top 20 rotation-function solutions gave the best solution with an R factor of 49.6% and a correlation coefficient of 37.2%, significantly above the next highest peak of R factor 53.9% and correlation coefficient 24.9%. Rigid-body refinement gave an R factor of 48.9% and a correlation coefficient of 42.8%. Crystal packing was good and initial isotropic temperature-factor refinement of the model produced an R factor of 32.5% and an R free of 42.7%. Improvements to the model were performed based on the electron density and the resulting structure was used as a search model to undertake molecular replacement again. Rotation-function calculations gave a peak (R factor 53.3%, correlation coefficient 26.7%) distinctly above the next highest peak (R factor 55.7%, correlation coefficient 19.1%). Translation-function calculations gave a solution (R factor 36.1%, correlation coefficient 68.9%) very significantly above the next symmetry-independent peak (R factor 54.8%, correlation coefficient 23.7%). Rigid-body refinement resulted in an R factor of 35.0% and a correlation coefficient of 72.8%. Structure refinement gave an R factor of 29.9% and an R free of 37.4%. Further improvement and refinement of the model is in progress. Currently, the electron density within the ligand binding-site is not clearly interpretable and is considered to arise from either a low-occupancy ligand or alternatively water molecules.
Figure 5.
Diffraction pattern from Wa VP8*64–223 crystal form A collected at the ESRF on a MAR CCD detector set at a distance of 175 mm. The edge of the image is 2.3 Å.
Table 1. Diffraction data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Form A, 100 K data | Form A, 295 K data | Form B, 100 K data | |
|---|---|---|---|
| Space group | P3221 | P3221 | P21 |
| Unit-cell parameters (Å, °) | a = b = 74.76, c = 70.09 | a = b = 76.00, c = 71.11 | a = 63.36, b = 160.32, c = 64.39, β = 119.28 |
| Crystal-to-detector distance (mm) | 175 | 65 | 190 |
| No. of images | 360 | 900 | 180 |
| Oscillation angle (°) | 0.5 | 0.2 | 1.0 |
| Exposure time (s) | 60 | 40 | 40 |
| Resolution (Å) | 24–2.5 (2.6–2.5) | 30–3.0 (3.14–3.0) | 26–2.8 (2.95–2.8) |
| Total No. of observations | 72933 (10494) | 18084 (1064) | 60111 (8138) |
| No. of unique reflections | 8135 (1097) | 4842 (619) | 24909 (2283) |
| Redundancy | 8.96 (9.6) | 3.7 (1.7) | 2.4 (3.6) |
| Completeness (%) | 97.4 (97.7) | 96.9 (98.7) | 78.3 (69.1) |
| Average I/σ(I) | 7.0 (1.9) | 9.0 (4.0) | 6.4 (3.6) |
| Rmerge† (%) | 9.7 (41.1) | 7.2 (15.8) | 10.7 (24.1) |
R
merge = 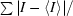
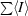 , where I is the intensity of each reflection.
, where I is the intensity of each reflection.
X-ray diffraction data (3.0 Å, Table 1 ▶) were collected at 295 K from a form A crystal grown in the absence of ligand and mounted inside a quartz capillary (Fig. 3 ▶ b). The Matthews coefficient V M is 3.2 Å3 Da−1 (solvent content 62%), assuming the presence of one molecule in the asymmetric unit of the trigonal system (space group P3221). Molecular replacement was performed using the Wa VP8*64–223 structure as determined from the crystal form A 100 K data. The highest rotation function peak had an R factor of 47.6% and a correlation coefficient of 30.7%; the next highest peak had an R factor of 48.6% and a correlation coefficient of 27.5%. Translation-function calculations gave a clear solution with an R factor of 30.5% and a correlation coefficient of 74.4%, significantly above the next peak (R factor 39.6%, correlation coefficient 51.8%). Rigid-body refinement produced an R factor of 29.4% and a correlation coefficient of 76.3%. Good crystal packing was apparent and refinement of the model is in progress.
X-ray diffraction data (2.8 Å, Table 1 ▶) were collected at 100 K from a form B crystal frozen in liquid nitrogen after being briefly dipped in 15%(v/v) glycerol and reservoir solution. The crystal exhibited a monoclinic system; however, it should be noted that the unit-cell parameters a and c are effectively equal and β is ∼120°. However, the alternative trigonal system with a = b = 63.29, c = 159.58 Å, γ = 120° did not give convincing data scaling-statistics. Here, we report the statistics associated with the structure solution derived within the monoclinic system (P21). The Matthews coefficient V M is 3.9, 3.1 and 2.6 Å3 Da−1 for four, five and six molecules, respectively, with associated solvent contents of 68, 61 and 45%, respectively. A molecular-replacement solution using the homology model gave a maximum of four molecules per asymmetric unit, with rigid-body refinement giving an R factor of 46.1% and a correlation coefficient of 45.9%. Initial refinement of the structure (R factor 34.3%, R free 41.4%) gave resulting electron-density with some ambiguities (although it should be noted that the completeness of the data is limited), raising concerns regarding the correctness of the solution. As the structure of Wa VP8*64–223 (crystal form A) became available this was used for molecular replacement and gave significantly improved discrimination amongst the potential rotation- and translation-function solutions. Four molecules were located and rigid-body refinement gave an R factor of 42.8% and a correlation coefficient of 51%. Molecular packing indicated possible accommodation of a fifth molecule, which a translation function (R factor 40.5%, correlation coefficient 55%; next highest peak, R factor 44%, correlation coefficient 46%) supported. Rigid-body refinement gave an R factor of 39.6%, a correlation coefficient of 58% and good molecular packing. Initial structure refinement (R factor 30.5%, R free 37.5%) gave reasonable electron density for the protein structure. The electron density within the carbohydrate-binding site is as yet uninterpretable, indicating either a weakly bound ligand or alternatively water molecules.
These studies represent the first report of structural data on VP8* from a rotavirus strain that infects humans and also the first relating to a sialidase-insensitive rotavirus strain. Having determined the three-dimensional structure of Wa VP8*64–223, it is anticipated that as this (and future) work progresses important insights and implications will be gained regarding interaction of this rotavirus spike protein with carbohydrate-based ligands and hence on aspects of host-cell specificity.
Acknowledgments
HB, BSC and MvI gratefully acknowledge the financial support of the Australian Research Council (ARC). HB thanks Dr Jean-Luc Ferrer for assistance in the operation of X-ray diffraction data-collection equipment at beamline FIP-BM30A (ESRF). Dr Alex Szyczew is acknowledged for synthesizing the methyl α-d-N-acetylneuraminide. BSC is supported by a Senior Research Fellowship from the National Health and Medical Research Council of Australia. MvI thanks the ARC for the award of an Australian Federation Fellowship.
References
- Ciarlet, M. & Estes, M. K. (1999). J. Gen. Virol.80, 943–948. [DOI] [PubMed] [Google Scholar]
- Ciarlet, M., Ludert, J. E., Iturriza-Gomara, M., Liprandi, F., Gray, J. J., Desselberger, U. & Estes, M. K. (2002). J. Virol.76, 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Coulson, B. S., Londrigan, S. H. & Lee, D. J. (1997). Proc. Natl Acad. Sci. USA, 94, 5389–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme, C., Brüssow, H., Sidoti, J., Roche, N., Karlsson, K.-A., Neeser, J.-R. & Teneberg, S. (2001). J. Virol.75, 2276–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer, P. R., Sun, Z.-Y. J., Wagner, G. & Harrison, S. C. (2002). EMBO J.21, 885–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith, M. L & Holmes, I. H. (1984). Nucleic Acids Res.12, 3973–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes, M. K. (2001). Fields Virology, 4th ed., edited by D. M. Knipe & P. M. Howley, p. 1747. Philadelphia: Lippincott–Raven Publishers.
- Estes, M. K. & Cohen, J. (1989). Microbiol. Rev.53, 410–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome, K., Yoshie, O. & Konno, T. (1989). Virology, 172, 196–205. [DOI] [PubMed] [Google Scholar]
- Guerrero, C. A., Zárate, S., Corkidi, G., López, S. & Arias, C. F. (2000). J. Virol.74, 9362–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, C. T., Nakagomi, O., Mochizuki, M., Ishida, H., Kiso, M., Ohta, Y., Suzuki, T. & Miyamoto, D. (1999). J. Biochem.126, 683–688. [DOI] [PubMed] [Google Scholar]
- Hewish, M. J., Takada, Y. & Coulson, B. S. (2000). J. Virol.74, 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Jolly, C. L., Beisner, B. M. & Holmes, I. H. (2000). Virology, 275, 89–97. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Parashar, U. D., Hummelman, E. G., Bresee, J. S., Miller, M. A. & Glass, R. I. (2003). Emerg. Infect. Dis.9, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, B. V. & Chiu, W. (1994). Curr. Top. Microbiol. Immunol.185, 9–29. [DOI] [PubMed] [Google Scholar]
- Sali, A. & Blundell, T. L. (1993). J. Mol. Biol.234, 779–815. [DOI] [PubMed] [Google Scholar]
- Santos, N. & Hoshino, Y. (2005). Rev. Med. Virol.15, 29–56. [DOI] [PubMed] [Google Scholar]
- Scott, S. A., Holloway, G., Coulson, B. S., Szyczew, A. J., Kiefel, M. J., von Itzstein, M. & Blanchard, H. (2005). Acta Cryst. F61, 617–620. [DOI] [PMC free article] [PubMed]