Recombinant rat cysteine dioxygenase (CDO) has been expressed, purified and crystallized and X-ray diffraction data have been collected to 1.5 Å resolution.
Keywords: cysteine, cysteine dioxygenase, sulfur amino acids, taurine, oxidative metabolism
Abstract
Cysteine dioxygenase (CDO; EC 1.13.11.20) is an ∼23 kDa non-heme iron metalloenzyme that is responsible for the oxidation of cysteine by O2, yielding cysteinesulfinate. CDO catalyzes the first step in the conversion of cysteine to taurine, as well as the first step in the catabolism of cysteine to pyruvate plus sulfate. Recombinant rat CDO was heterologously expressed, purified and crystallized. The protein was expressed as a fusion protein bearing a polyhistidine tag to facilitate purification, a thioredoxin tag to improve solubility and a factor Xa cleavage site to permit removal of the entire N-terminus, leaving only the 200 amino acids inherent to the native protein. A multi-step purification scheme was used to achieve >95% purity of CDO. The optimal CDO crystals diffracted to 1.5 Å resolution and belonged to space group P43212 or P41212, with unit-cell parameters a = b = 57.55, c = 123.06 Å, α = β = γ = 90°. CDO shows little homology to any other proteins; therefore, the structure of the enzyme will be determined by ab initio phasing using a selenomethionyl derivative.
1. Introduction
Cysteine dioxygenase (CDO; EC 1.13.11.20) is a mononuclear non-heme iron protein that catalyzes the addition of molecular oxygen to the thiol group of cysteine to form cysteinesulfinate. CDO shows little sequence similarity with any other protein, but does appear to be widespread in eukaryotes and eubacteria. Within mammals, CDO mRNA is expressed in a tissue-specific manner. Usually, CDO activity is much more abundant in the liver than in any other tissue, but appreciable levels of CDO activity are also found in the kidney, lung and brain (Hirschberger et al., 2001 ▶; Tsuboyama et al., 1996 ▶; Shimada et al., 1998 ▶; Stipanuk et al., 2002 ▶).
By catalyzing the first step in the oxidative metabolism of cysteine, CDO plays a key role in cysteine catabolism, in the provision of cysteine carbon (pyruvate) for gluconeogenesis or oxidative metabolism, in taurine biosynthesis and in the supply of inorganic sulfate for sulfation reactions. The end products of the pathways of cysteine metabolism that are initiated by CDO, taurine and sulfate, are essential themselves for bile-acid conjugation, osmotic regulation, synthesis of glycosaminoglycans, sulfation of xenobiotics and other diverse functions.
Although no polymorphism of the human CDO gene has been described, clinical findings of depressed levels of sulfate in plasma, elevated fasting plasma cysteine concentrations and elevated plasma cysteine:sulfate ratios in individuals with rheumatoid arthritis, liver disease, Parkinson’s disease, Alzheimer’s disease, motor neuron disease and systemic lupus erythematosus suggest that low levels of functional CDO may contribute to human disease (Bradley et al., 1994 ▶; Davies et al., 1995 ▶; Heafield et al., 1990 ▶, 1992 ▶). Patients in these groups displayed an impaired capacity for sulfation reactions in vivo. Thus, an insufficient supply of sulfate or taurine and impairment of sulfation reactions could be contributory factors in the etiology of some of these diseases. Additionally, elevated levels of cysteine could contribute to the etiology of any of these diseases.
We now report the expression and purification of recombinant rat liver cysteine dioxygenase. Using highly purified CDO, we determined optimal conditions for CDO crystallization and obtained crystals that were suitable for X-ray diffraction analysis. The reported purification scheme and crystallographic characterization are critical steps towards elucidating a structural model for CDO and probing the catalytic mechanism of this distinctive dioxygenase.
2. Methods
2.1. Cloning and expression
First-strand rat liver CDO cDNA was prepared via RT-PCR with the Superscript First Strand Synthesis System (Invitrogen). Primers were designed for use with the ligation-independent cloning (LIC) vector pET-30 Xa/LIC (Novagen) using forward primer 5′-GGTATTGAGGGTCGCATGGAACGGACCGAGCTGCTG-3′ and reverse primer 3′-AGAGGAGAGTTAGAGCCCTATTAGTTGTTCTCCAGTGAACCTGAAG-5′. The PCR product was inserted into the vector using the Xa/LIC cloning kit. The construct was then used to transform Nova Blue Escherichia coli (Novagen). A portion of this construct was subsequently cloned into the pET32a vector using the BglII and NcoI restriction sites found on both plasmids, yielding a pET32a construct with a factor Xa cleavage site just before the CDO-ORF rather than the enterokinase site normally found on that plasmid. Vector pET32a contains a thioredoxin protein sequence 5′ to the CDO-ORF, causing a fusion protein to be formed. It also contains a 6×His tag between the thioredoxin sequence and the factor Xa site. The sequence of the construct in the pET32a vector was verified at the Cornell University BioResource Center. The rCDO/pET32a vector was then transformed into E. coli strain BL21(DE3) competent cells (Novagen) for expression of the rCDO fusion protein. For protein expression, cells were grown in 2× Luria Broth (LB) at 310 K containing 100 µg ml−1 carbenicillin. Cells were grown to an OD600 of ∼0.6, at which point expression of rCDO was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cells were grown for an additional 3 h at 310 K and then harvested by centrifugation.
2.2. Purification
The cells harvested from 3 l culture medium were resuspended in 50 ml lysis buffer containing 20 mM Tris pH 8.0, 5 mM imidazole, 0.1%(v/v) Tween, 500 mM sodium chloride and one tablet of Complete protease inhibitor (Roche). Cells were lysed via sonication with an Ultrasonic Sonicator (Misonix) and then centrifuged at 30 000g for 30 min to remove cellular debris. The supernatant was filtered with a 0.2 µm syringe filter and filtered lysate was applied onto a 1 ml HisTrap HP column (Amersham Biosciences). The protein was chromatographed at a flow rate of 1 ml min−1 by generating increasing imidazole concentrations with 20 mM Tris pH 8.0, 5 mM imidazole, 0.1%(v/v) Tween, 500 mM sodium chloride (IMAC buffer A) mixed with 20 mM Tris pH 8.0, 500 mM imidazole, 0.1%(v/v) Tween and 500 mM sodium chloride (IMAC buffer B). CDO peak fractions that eluted with 50 mM imidazole were recovered, concentrated and then diluted with 20 mM Tris pH 8.0 with 0.1% Tween (IEX buffer A) to dilute the salt concentration to <20 mM. The sample was then loaded onto a MonoQ 4.6/100 PE ion-exchange column (Amersham Biosciences) and chromatographed at 1 ml min−1 using a linear NaCl gradient generated with 20 mM Tris pH 8.0 with 0.1% Tween (IEX buffer A) and 20 mM Tris pH 8.0, 0.1% Tween and 1 M NaCl (IEX buffer B). The linear gradient from 0 to 500 mM NaCl was generated over 30 CVs (1 CV is ∼2 ml). Peak fractions were pooled and concentrated to <2 ml and applied onto a Superdex 200 16/60 column (Amersham Biosciences) equilibrated with 20 mM Tris pH 8.0, 100 mM NaCl at a flow rate of 1 ml min−1. Peak fractions were recovered and then incubated in the presence of 0.3 U µl−1 factor Xa (Novagen) overnight at 277 K to remove the thioredoxin-6×His fusion tag. The following day, the factor Xa protease was removed from the reaction solution using Xarrest agarose (Novagen). The solution was again run over the 1 ml HisTrap HP column to bind and remove undigested protein as well as the thioredoxin-6×His N-terminal domain. Flowthrough fractions from the column were collected and the protein concentration was determined using A 280 and an extinction coefficient of 25 230 cm−1 M −1. The overall yield was 1–2 mg of pure CDO per litre of culture. NuPAGE 4–12% SDS–PAGE gels (Invitrogen) were run and stained with Coomassie blue at each step of the purification to estimate protein purity and abundance. The molecular weight of purified CDO as determined by MALDI–TOF at the Cornell BioResource Center was 23 033 ±10 Da and was in good agreement with the theoretical weight of CDO predicted based upon the primary sequence (23 026 Da).
3. Results and discussion
3.1. Crystallization
Native CDO (200 amino acids, ∼23 kDa) and full-length fusion protein (∼40.3 kDa), each purified to homogeneity, were subjected to extensive crystallization screening. Purified CDO was screened for crystal formation at several protein concentrations with Crystal Screens 1 and 2 from Hampton Research using the sitting-drop vapour-diffusion method. 1 µl reservoir solution was mixed with 1 µl protein solution in each drop and the crystals were all grown at room temperature. CDO purified using an original purification scheme without the IEX step was also screened. No crystals were observed in any of the crystallization drops containing the full-length fusion protein or containing CDO that had been purified by the initial purification scheme that did not include the IEX step. Overloaded SDS–PAGE gels from the protein purified using the original purification scheme showed the presence of a minor contaminant co-purifying with CDO, leading to the conclusion that this could perhaps be preventing crystallization. However, by including the IEX step in the purification scheme and then removing the large fusion tag, we obtained a homogeneous and soluble preparation of CDO that readily crystallized under multiple conditions. The fact that only the highly purified CDO crystallized accentuates the importance of protein purity for successful crystallization.
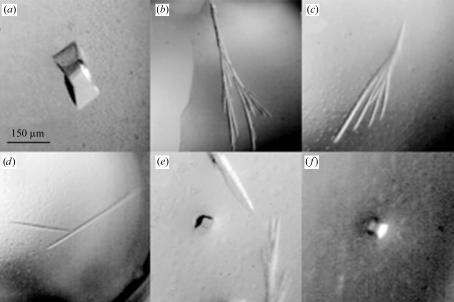
The best crystals were observed in Hampton Research Crystal Screen Kit 1 condition No. 9 [0.2 M ammonium acetate plus 0.1 M trisodium citrate pH 5.6, with 30%(w/v) polyethylene glycol 4000 (PEG 4K)] with a protein concentration of 7.5 mg ml−1. Subsequent screening was performed by varying the PEG 4K and ammonium acetate concentrations; optimal crystals appeared in drops containing 22–26%(w/v) PEG 4K and 0.1–0.25 M ammonium acetate. Multiple crystal forms appeared in a variety of precipitant concentrations in the optimization screens around the original condition. These included long needles with dimensions of 0.75 × 0.05 mm (Figs. 1 ▶ b, 1 ▶ c and 1 ▶ d), a crystal form resembling an arrowhead growing to 0.15 × 0.05 mm (Fig. 1 ▶ e) and cubes that were 0.05–0.1 mm in each dimension when fully grown (Figs. 1 ▶ e and 1 ▶ f). Apart from the cube-shaped crystals, the other crystals had a propensity for multiple crystal growth from a single point of nucleation (Figs. 1 ▶ b and 1 ▶ c) and were very often observed to be polycrystalline (Fig. 1 ▶ a).
Figure 1.
CDO crystallizes in a variety of crystal forms. (a): example of a twinned crystal which was observed in a number of drops under a variety of conditions. X-ray diffraction analysis of this polycrystalline crystal form confirmed the presence of multiple lattices (data not shown). (b), (c) and (d): examples of the most common CDO crystal morphologies. Needles very often formed in drops in tree-shaped (b) or fan-shaped (c) structures originating from single points of nucleation and at times were also found as long thin single crystals (d). (e) and (f): the cube-shaped morphology shown here yielded the highest resolution diffraction data, including the 1.5 Å data outlined in this report.
3.2. Data collection and processing
The data described in this report were collected from a cube-shaped crystal that was crystallized in an optimization screen of Hampton Research Crystal Screen condition No. 9. The crystal grew to ∼0.1 mm in each dimension in 10 d at room temperature in a solution containing 0.1 M ammonium acetate plus 0.1 M trisodium citrate pH 5.6 with 26%(w/v) PEG 4K and a CDO concentration of 7.5 mg ml−1. Each of the crystal-containing drops contained solution that was deemed to be sufficient for proper cryoprotection and prolonged X-ray exposure during data collection. Cryoloops (Hampton Research) were used for mounting CDO crystals, which were frozen by plunging into a liquid-nitrogen bath or via direct insertion into the nitrogen cold stream. Collection of crystallographic data from CDO crystals was performed at the National Synchrotron Light Source (NSLS) X12b beamline on an ADSC Q4 CCD detector at λ = 1.0 Å at 100 K. A 1.5 Å native data set consisting of 180 frames was collected with a crystal-to-detector distance of 150 mm, an oscillation angle of 1° and an exposure time of 20 s per frame. The X-ray diffraction data were processed and scaled using HKL2000 (Otwinowski & Minor, 1997 ▶). The CDO crystals were found to belong to space group P43212 or P41212, with unit-cell parameters a = b = 57.55, c = 123.06 Å, α = β = γ = 90°. The data set was 98.2% complete to 1.5 Å. The crystallographic data statistics are summarized in Table 1 ▶. The Matthews coefficient was 2.3 Å3 Da−1, indicating a solvent content of 45.2% assuming the presence of one molecule of CDO in the asymmetric unit (Matthews, 1968 ▶). Multiple crystal lattices were observed in the X-ray diffraction patterns of a significant number of the crystals, primarily those with the needle morphology; however, the cube-shaped crystal forms were observed to be single crystals and yielded the best diffraction data.
Table 1. Crystallographic data statistics.
Values in parentheses are for the highest resolution shell.
| Experimental station | NSLS X12b |
| X-ray wavelength (Å) | 1.0 |
| Unit-cell parameters (Å) | a = b = 57.55, c = 123.06 |
| Space group | P43212 or P41212 |
| Resolution (Å) | 30–1.50 (1.55–1.50) |
| Unique reflections | 33453 |
| Multiplicity | 13.1 |
| I/σ(I) | 52.4 (7.0) |
| Rsym (%) | 4.6 (31.8) |
| Completeness (%) | 98.2 (95.8) |
The structure of the enzyme will be determined by ab initio phasing using a selenomethionyl-substituted derivative of CDO and attempts at this have already begun. The purification scheme and crystallization conditions that we report will greatly enhance our ability to elucidate the crystal structure of CDO and will significantly improve our ability to study the catalytic mechanism of this important enzyme.
Acknowledgments
We would like to thank Dr Qingqiu Huang for technical support and discussion regarding CDO crystallization experiments and Mr Robert Sherwood at the Cornell BioResoure Center for performing the mass-spectral analysis. We would also like to acknowledge Dr Dieter Schneider and Dr Alexei Soares at the NSLS X12b beamline for their technical support. This work was supported by National Institutes of Health grant PHS DK056649 awarded to MHS. Support for the crystallization experiments was funded by award RR-01646 from the National Institutes of Health through its National Center for Research Resources. Financial support for beamline X12b of the NSLS comes principally from the Offices of Biological and Environmental Research, Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources of the National Institutes of Health.
References
- Bradley, H., Gough, A., Sokhi, R. S., Hassell, A., Waring, R. & Emery, P. (1994). J. Rheumatol.21, 1192–1196. [PubMed] [Google Scholar]
- Davies, M. H., Ngong, J. M., Pean, A., Vickers, C. R., Waring, R. H. & Elias, E. (1995). J. Hepatol.22, 551–560. [DOI] [PubMed] [Google Scholar]
- Heafield, M. T., Fearn, S., Steventon, G. B., Waring, R. H., Williams, A. C. & Sturman, S. G. (1990). Neurosci. Lett.110, 216–220. [DOI] [PubMed] [Google Scholar]
- Heafield, M. T. & Williams, A. C. (1992). Curr. Opin. Neurol. Neurosurg.5, 288–294. [PubMed] [Google Scholar]
- Hirschberger, L. L., Daval, S., Stover, P. J. & Stipanuk, M. H. (2001). Gene, 277, 153–161. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shimada, M., Koide, T., Kuroda, E., Tsuboyama, N., Hosokawa, Y. & Watanabe, M. (1998). Amino Acids, 15, 143–150. [DOI] [PubMed] [Google Scholar]
- Stipanuk, M. H., Londono, M., Lee, J. I., Hu, M. & Yu, A. F. (2002). J. Nutr.132, 3369–3378. [DOI] [PubMed] [Google Scholar]
- Tsuboyama, N., Hosokawa, Y., Totani, M., Oka, J., Matsumoto, A., Koide, T. & Kodama, H. (1996). Gene, 181, 161–165. [DOI] [PubMed] [Google Scholar]