Crystals of the type I Baeyer–Villiger monooxygenase (BVMO) MtmOIV from the biosynthetic pathway of mithramycin were obtained; the crystals diffracted to 2.69 Å resolution and belong to the monoclinic space group C2 (a = 143.5, b = 114.2, c = 137.8 Å β = 102.5°). Light scattering indicates that MtmOIV is a dimer of 127 kDa in solution, while in the crystalline state the data are consistent with two dimers in the asymmetric unit.
Keywords: mithramycin, MtmOIV, Baeyer–Villiger monooxygenase, FAD-dependent, NADPH-dependent
Abstract
The Baeyer–Villiger monooxygenase MtmOIV from Streptomyces argillaceus is a 56 kDa FAD-dependent and NADPH-dependent enzyme that is responsible for the key frame-modifying step in the biosynthesis of the natural product mithramycin. Crystals of MtmOIV were flash-cooled and diffracted to 2.69 Å resolution using synchrotron radiation on beamline SER-CAT 22-ID at the Advanced Photon Source. Crystals of MtmOIV are monoclinic and light-scattering data reveal that the enzyme forms dimers in solution. The rotation function suggests the presence of two dimers in the asymmetric unit. l-Selenomethionine-incorporated MtmOIV has been obtained. Structural solution combining molecular-replacement phases and anomalous phases from selenium is in progress.
1. Introduction
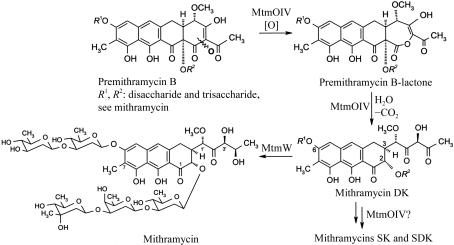
Mithramycin (MTM; also known as aureolic acid, mithracin, LA-7017, PA-144 and plicamycin) is an aureolic acid-type polyketide anticancer antibiotic produced by the soil bacterium Streptomyces argillaceus (ATCC 12956; Remsing et al., 2003 ▶; Prado et al., 1999 ▶; Rohr et al., 1999 ▶). MTM displays anticancer activity through its ability to inhibit replication and transcription via cross-linking of DNA strands (Gao & Patel, 1990 ▶). Combinatorial biosynthesis, in which the genes of the biosynthetic pathway are altered in the bacteria themselves (e.g. through gene inactivation, overexpression or recombination), has been used to generate new mithramycin analogues with an improved therapeutic index (Remsing et al., 2003 ▶; Albertini et al., 2005 ▶). This method not only allows the generation of new analogs, so-called ‘hybrid products’ or ‘unnatural natural compounds’, but also simultaneously generates a bacterial strain which allows the biotechnological mass production of the new compound. MtmOIV is responsible for a series of reactions leading to an oxidative C—C bond cleavage (Prado et al., 1999 ▶; Gibson et al., 2005 ▶; Rodriguez et al., 2003 ▶), which essentially establishes the characteristic molecular frame found in all aureolic acid drugs and converts biologically completely inactive premithramycin B into active molecules, initially mithramycin DK (Fig. 1 ▶) (Albertini et al., 2005 ▶; Gibson et al., 2005 ▶). Although this enzyme revealed some degree of substrate flexibility, many of the previously generated premithramycins, particularly those with incomplete or altered saccharide moieties, could not be processed at all by MtmOIV or could only be processed insufficiently (Prado et al., 1999 ▶; Nur-e-Alam et al., 2005 ▶; Remsing et al., 2002 ▶, 2003 ▶). Knowledge of the crystal structure of MtmOIV will allow the structure-based engineering of this enzyme for the purpose of broadening its substrate specificity to allow the generation of many more and much more chemically diverse mithramycin analogs.
Figure 1.
Reactions involving MtmOIV.
Baeyer–Villiger monooxygenases (BVMOs) catalyze the insertion of an O atom next to a keto function to yield an ester or a lactone. Their mechanism is conceptually identical to the non-enzymatic Baeyer–Villiger rearrangement reaction, except that FAD-bound dioxygen as opposed to synthetic peroxides initiates the reaction. The function of MtmOIV as a Baeyer–Villiger monooxygenase (BVMO) has already been suggested from gene-deletion experiments and studies with 18O-labeled precursors (Prado et al., 1999 ▶; Rohr et al., 1999 ▶; Rodriguez et al., 2003 ▶) and has recently been proven through studies using the overexpressed enzyme (Gibson et al., 2005 ▶). The latter investigations revealed that the expected Baeyer–Villiger rearrangement lactone product, premithramycin BL, is indeed the immediate product of the reaction of MtmOIV with its substrate premithramycin B and is the first intermediate of the reaction cascade catalyzed by this unusual oxygenase (Fig. 1 ▶). While many organisms have been reported to produce enzymes that are capable of catalyzing Baeyer–Villiger reactions on artificial substrates and thus have potential for use as biocatalysts in organic synthesis (Kamerbeek et al., 2003 ▶; Alphand et al., 2003 ▶; Gutierrez et al., 2003 ▶), it is important in the context of this communication to point out that MtmOIV is the only BVMO so far that has been characterized as such with its natural substrate. Thus, in contrast to other described BVMOs, which were found to catalyze Baeyer–Villiger rearrangement reactions when confronted with small synthetic cyclic or aliphatic ketones while their natural substrates and reaction mechanisms remain obscure, MtmOIV performs a Baeyer–Villiger reaction with its natural substrate, premithramycin B, a comparably large molecule. Although MtmOIV carries structural features that have previously been implicated in typical type I BVMOs, such as an aspartate located five residues upstream of the binding motif for the adenosine moiety of FAD (Willetts, 1997 ▶) or a second βαβ or Rossmann-fold (GxGxxG) motif (Kamerbeek et al., 2004 ▶), MtmOIV appears to belong to a different, possibly new, class of FAD-dependent and NADPH-dependent BVMOs and is rather unrelated to the recently structurally characterized type I BVMO phenylacetone monooxygenase (PAMO) from Thermobifida fusca (Malito et al., 2004 ▶).
2. Experimental methods and results
2.1. Cloning and purification of and l-selenomethionine incorporation into MtmOIV
The mtmOIV gene was cloned by PCR using pLPO4 as a template (Prado et al., 1999 ▶) and the advantage-GC polymerase mix (BD Bioscience Clonetech). The forward primer 5′-CGCACGGATCCCGGAAGGCGCATGATG-3′ and the reverse primer 5′-ATCGTCCAGGTTGTGAATTCGGGCGCCGA-3′ were designed to incorporate BamHI and EcoRI restriction sites (in bold). The 1.7 kbp mtmOIV PCR product, double digested, was ligated into the BamHI and EcoRI sites of the N-terminal His6-tag expression vector pRSETb with T4 DNA ligase. The recombinant plasmid was re-isolated and confirmed by DNA-sequencing and restriction analysis. The expression vector was transformed into Escherichia coli strain BL21(DE3)pLysS. A 1 l culture of LB medium containing 1 mM ampicillin and 1 mM chloramphenicol was inoculated with 50 ml of an overnight-grown culture and incubated at 310 K until the OD at 600 nm reached 0.5–0.7. IPTG was added to a final concentration of 1 mM and the culture was grown at 291–293 K for an additional 5 h. Cells were harvested by centrifugation at 4000g for 20 min and stored at 193 K.
The cell pellet was resuspended in 50 ml ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl pH 8.0) and disrupted by lysozyme (1 mg ml−1) and ultrasonic treatment for 6 × 10 s. After lysis, 1%(w/w) protamine sulfate was added and the homogenate was then clarified by centrifugation at 10 000g at 279 K for 30 min. The supernatant was loaded onto a 2 ml Sepharose-based Talon metal-affinity resin column equilibrated with equilibration buffer (50 mM NaH2PO4, 300 mM NaCl pH 7.0). The enzyme was eluted using elution buffer (150 mM imidazole in equilibration buffer). The purification yield was 3–4 mg per litre of culture. SDS–PAGE showed a single band at around 60 kDa which is in good agreement with a calculated subunit molecular weight of 56.1 kDa. To express SeMet-lablled protein, the mtmOIV::pRSETb vector was transformed into a competent E. coli B834(DE3) host strain. Incorporation followed the procedure described by Ramakrishnan & Biou (1997 ▶).
2.2. Crystallization
The freshly purified MtmOIV was buffer-exchanged and concentrated to 6–7 mg ml−1 using a Centricon Plus-20 (Amicon) in crystallization buffer (0.1 M Tris–HCl pH 7.6–8.0, 10 µM FAD, 0.1% NaN3, 0.5 mM PMSF). The initial crystallization trials were carried out using the vapor-diffusion technique with commercial kits (Structure Screen 2, Molecular Dimensions Inc.). The hanging drops consisted of 4 µl MtmOIV solution and 4 µl reservoir solution. Crystals grew using reservoir solution A [0.1 M Na HEPES pH 7.5, 10%(v/w) PEG 6000 and 5%(v/v) MPD, condition No. 12] or reservoir solution B [0.1 M Na HEPES pH 7.5, 10%(v/w) PEG 8000 and 8%(v/v) ethylene glycol, condition No. 19]. Crystals were grown at room temperature and reached dimensions of 0.2–0.3 mm in 3 d. Optimization of the crystallization conditions was performed by altering both the MPD and ethylene glycol concentrations. Plate-shaped crystals (up to 0.5 mm in the largest dimension) grew in reservoir solutions containing 5–14% MPD or 8–16% ethylene glycol. Smaller hexahedral crystals were occasionally observed when the concentration of MPD was increased to about 15–16%. When the temperature was 277 K, only showers of crystals were formed.
2.3. Dynamic light scattering and oligomeric state
Experiments were performed using MtmOIV (46.4 µM) with bound FAD. The MtmOIV solution had a mean polydispersity of 0.97 and a monomodal fit of the data indicated a molecular weight of 127.6 kDa, consistent with the presence of a dimer in solution at pH 8.0. The polydispersity index suggested conditions suitable for crystallization. Dynamic light-scattering measurements were performed using a DynaPRO99 instrument (Protein Solutions). MtmOIV was concentrated to approximately 2.6 mg ml−1 in crystallization buffer. The protein solution was filtered through 0.1 µm Anodisc 13 membrane filters (Whatman) to remove any residual gas bubbles or solid impurities. Dynamics v.5.25.44 software was used in the data collection and analysis. The number of acquisition scans was 20 and the acquisition time was 5 s per scan at 295 K.
2.4. X-ray data collection and analysis
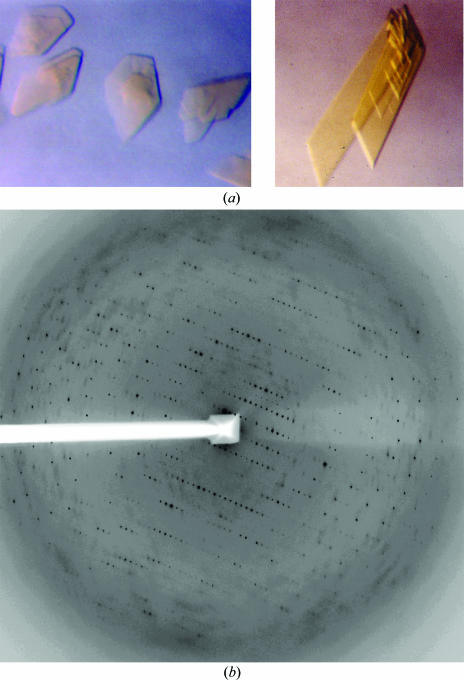
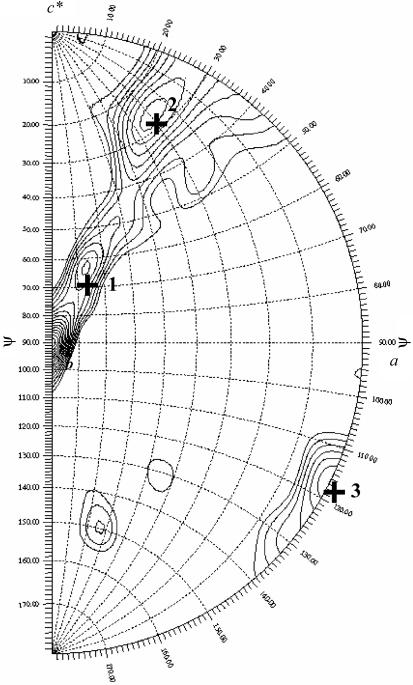
Crystals were harvested directly from hanging drops using Hampton cryoloops and flash-cooled in liquid nitrogen. We were able to collect a near-complete data set from a single crystal at the SER-CAT 22-ID beamline at APS-Argonne, IL, USA (Table 1 ▶). Crystals diffracted to 2.69 Å at the synchrotron source and to below 4 Å using a standard rotating-anode source. More than one crystal is necessary for data completeness. Data from two crystals were processed to 2.8 Å using HKL2000 (Otwinowski & Minor, 1997 ▶) (Table 1 ▶). A typical diffraction pattern is shown in Fig. 2 ▶. Given the unit-cell parameters of MtmOIV crystals (very little variation in unit-cell parameters was observed between crystals) and the molecular weight of the dimer, it can be estimated that crystals of MtmOIV have two dimers in the asymmetric unit. A calculation of a self-rotation function using the program GLRF (Tong & Rossmann, 1997 ▶) allows identification of the molecular twofold axis. A stereographic projection of the rotation function for κ = 180 (twofold peaks) is shown in Fig. 3 ▶. The largest peak (ϕ = 90, ψ = 0°) corresponds to the crystallographic twofold axis of the space-group symmetry C2. There are three twofold non-crystallographic peaks in the rotation function, as indicated in Fig. 3 ▶ with the symbol +. Peak 3 at ϕ = 0 and ψ = 117° is roughly perpendicular to the other two twofold axis (2 and 1 in Fig. 3 ▶). All three peaks are quite close to a 222 point-group arrangement. The native Patterson map does not indicate any strong peak consistent with a translation between dimers. The non-crystallographic peaks are consistent with the presence of two dimers in the asymmetric unit. Selenium-incorporated crystals displayed a larger degree of radiation decay and lower resolution was observed. Based on sequence similarity (25% identity to phenol hydroxylase, 8% identity to PAMO) and a significantly different fingerprint (region 167–177 in PAMO), MtmOIV possibly belongs to an uncharacterized class of Baeyer–Villiger monooxygenases that is part of the GR2 class of FAD-dependent enzymes (Dym & Eisenberg, 2001 ▶). An increase in diffraction data completeness to 2.6 Å can be obtained with larger crystals, as they allow more data to be collected from a single crystal. Our strategy for structural solution will rely on combining both molecular replacement and Se data.
Table 1. Crystallographic parameters of MtmOIV crystals and data-collection statistics.
| Native | Se edge | Se peak | |
|---|---|---|---|
| Unit-cell parameters | |||
| a (Å) | 143.5 | ||
| b (Å) | 114.2 | ||
| c (Å) | 137.8 | ||
| V (Å3) | 1343663 | ||
| Space group | C2 | ||
| Monomer MW (kDa) | 56 | ||
| Dimers in ASU | 2 | ||
| VM (Å3 Da−1) | 2.85 | ||
| Redundancy | 3.6 | 5.9 | 7.8 |
| Resolution (Å) | 50–2.8 | 50–3.5 | 50–3.63 |
| Wavelength (Å) | 1.0 | 0.97942 | 0.97922 |
| No. of crystals | 2 | 2 | 1 |
| Beamline | 22-ID, APS | 22-ID, APS | 22-ID, APS |
| Rmerge (%) | 12.6 | 8.7 | 9.7 |
| Completeness (%) | 81.9 | 98.4 | 89.4 |
| I/σ(I) | 6.4 | 9.9 | 7.8 |
Figure 2.
(a) Crystals of MtmOIV. Left, native MtmOIV; right, Se-MtmOIV. Both crystals are flat plates and their largest dimension is 0.3–0.5 mm. (b) Representative diffraction pattern of native MtmOIV crystals collected on the SER-CAT 22-ID beamline at APS-Argonne, IL, USA using a MAR 300 detector with a crystal-to-detector distance of 320 mm.
Figure 3.
Rotation function, κ = 180. The data are consistent with two dimers in the asymmetric unit.
Acknowledgments
Major support of this research was provided by the National Institutes of Health (NIH grant CA 091901) to JR; Kentucky Lung Cancer Research Foundation grants supporting MAO and JR partially supported this research. Finally, we thank the SER-CAT staff for help during data collection. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. The University of Kentucky is a member institution of SER-CAT. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Albertini, V., Vignati, S., Rinaldi, A., Nur-e-Alam, M., Kwee, I., Riva, C., Donata Micello, D., Capella, C., Bertoni, F., Carbone, G. M., Rohr, J. & Catapano, C. V. (2005). Submitted.
- Alphand, V., Carrea, G., Wohlgemuth, R., Furstoss, R. & Woodley, J. M. (2003). Trends Biotechnol.21, 318–323. [DOI] [PubMed] [Google Scholar]
- Dym, O. & Eisenberg, D. (2001). Protein Sci.10, 1712–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. & Patel, D. J. (1990). Biochemistry, 29, 10940–10956. [DOI] [PubMed] [Google Scholar]
- Gibson, M., Nur-e-Alam, M., Lipata, F., Oliveira, M. A. & Rohr, J. (2005). Submitted. [DOI] [PubMed]
- Gutierrez, M. C., Sleegers, A., Simpson, H. D., Alphand, V. & Furstoss, R. (2003). Org. Biomol. Chem.1, 3500–3506. [DOI] [PubMed] [Google Scholar]
- Kamerbeek, N. M., Fraaije, M. W. & Janssen, D. B. (2004). Eur. J. Biochem.271, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Kamerbeek, N. M., Janssen, D. B., van Berkel, W. J. H. & Fraaije, M. W. (2003). Adv. Synth. Catal.345, 667–678.
- Malito, E., Alfieri, A., Fraaije, M. W. & Mattevi, A. (2004). Proc. Natl Acad. Sci. USA, 101, 13157–13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur-e-Alam, M., Mendez, C., Salas, J. A. & Rohr, J. (2005). Chembiochem, 6, 632–636. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Prado, L., Fernandez, E., Weissbach, U., Blanco, G., Quiros, L. M., Brana, A. F., Mendez, C., Rohr, J. & Salas, J. A. (1999). Chem. Biol.6, 19–30. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. & Biou, V. (1997). Methods Enzymol.276, 538–557. [PubMed] [Google Scholar]
- Remsing, L. L., Garcia-Bernardo, J., Gonzalez, A. M., Kunzel, E., Rix, U., Brana, A. F., Bearden, D. W., Mendez, C., Salas, J. A. & Rohr, J. (2002). J. Am. Chem. Soc.124, 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsing, L. L., Gonzalez, A. M., Nur-e-Alam, M., Fernandez-Lozano, M. J., Brana, A. F., Rix, U., Oliveira, M. A., Mendez, C., Salas, J. A. & Rohr, J. (2003). J. Am. Chem. Soc125, 5745–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, D., Quiros, L. M., Brana, A. F. & Salas, J. A. (2003). J. Bacteriol.185, 3962–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr, J., Mendez, C. & Salas, J. A. (1999). Bioorg. Chem.27, 41–54.
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]
- Willetts, A. (1997). Trends Biotechnol.15, 55–62. [DOI] [PubMed] [Google Scholar]