Nucleotide-exchange factor from S. solfataricus (SsEF-1β) has been successfully crystallized. X-ray diffraction data have been collected from the native enzyme and from the selenomethionine derivative of SsEF-1β to 1.97 and 1.83 Å resolution, respectively.
Keywords: protein biosynthesis, nucleotide-exchange factor, Sulfolobus solfataricus
Abstract
The nucleotide-exchange factor isolated from the hyperthermophilic archaeon Sulfolobus solfataricus (SsEF-1β) consists of 90 residues and differs from eukaryal EF-1βs. The protein has been successfully crystallized using either microbatch-under-oil or vapour-diffusion methods. Crystals of native SsEF-1β diffract to 1.97 Å resolution and belong to space group P21212, with unit-cell parameters a = 106.46, b = 54.87, c = 44.03 Å. Diffraction data have also been collected from a selenomethionine derivative of SsEF-1β at 1.83 Å resolution. Model building using the phases derived from the MAD experiment is in progress.
1. Introduction
Protein biosynthesis is a key event in all living organisms. Most of the actions linked to this biological process are performed by the ribosome (Ramakrishnan, 2002 ▶). However, ribosome function relies on several ancillary proteins such as initiation, elongation and termination factors. A particularly important role is played by the elongation factors EF-Tu/EF-1α that carry the aminoacyl-tRNA to the ribosome in the elongation cycle of the process (Andersen et al., 2003 ▶). These elongation factors are GTPases characterized by two important specific properties: (i) an extremely low intrinsic GTPase activity and (ii) a high affinity for the reaction product (GDP). The affinity for GDP is generally higher than that observed for the substrate (GTP). These features allow the transport of the aminoacyl-tRNA, bound to the GTP complex of EF-Tu/EF-1α, to the ribosome, as well as efficient modulation of the elongation factor function. The GDP is so tightly bound to these GTPases that other accessory proteins, denoted nucleotide-exchange factors, are necessary for GDP release and for regeneration of the active GTP-bound form of these enzymes. In contrast to elongation factors, which display some common basic properties in all forms of life, exchange factors isolated from the three domains of life display larger differences (Cherfils & Chardin, 1999 ▶). Indeed, no sequence similarity is detected between exchange factors isolated from bacteria (EF-Ts) and those extracted from archaea/eukarya (EF-1βs). Moreover, differences are also observed between archaeal and eukaryal EF-1βs. The molecular mass of eukaryal EF-1βs (23 kDa) is twice that exhibited by archaeal counterparts. A limited sequence identity (approximately 20%) is observed between the C-terminal domain of eukaryal exchange factors and archaeal EF-1βs.
Structural studies have been carried out by NMR on human (C-terminal fragment; Perez et al., 1999 ▶) and Methanobacterium thermoautotrophicum EF-1β (MtEF-1β; Kozlov et al., 2000 ▶). The crystallographic structure of the C-terminal fragment of Saccharomyces cerevisiae EF-1β in complex with the corresponding elongation factor (ScEF-1α–EF-1β) has also been reported (Andersen et al., 2000 ▶, 2001 ▶).
The exchange factor EF-1β isolated from the hyperthermophilic archaeon Sulfolobus solfataricus (SsEF-1β) has some distinctive properties (Arcari et al., 1995 ▶; Raimo et al., 1996 ▶). This protein is endowed with a remarkable thermostability. Indeed, no transition is observed in thermal denaturation curves up to 378 K monitored by far-UV circular dichroism (unpublished results) and 10 h of heating at 373 K is required to achieve 90% inactivation of the protein (Raimo et al., 1996 ▶). Additionally, non-denaturing gel filtration of SsEF-1β indicates an apparent molecular mass of 20 kDa, suggesting a dimeric form of this exchange factor (Raimo et al., 1996 ▶), since the protein consists of 90 residues with a corresponding molecular mass of 10 kDa. Furthermore, solution studies of the complex between SsEF-1β and its elongation-factor counterpart (SsEF-1α) have also shown an unexpected stoichiometry that indicates that two monomers of the exchange factor bind to a single elongation-factor molecule (Raimo et al., 1999 ▶). Therefore, structural studies on SsEF-1β are important for elucidation of its structure–stability and structure–function relationships. Here, we report the crystallization and preliminary crystallographic investigation of this protein.
2. Experimental methods
2.1. Purification and crystallization
Recombinant SsEF-1β was expressed in Escherichia coli BL21(DE3) strain as previously reported (Ianniciello et al., 1998 ▶). E. coli cells were mechanically disrupted and the cell debris was removed by centrifugation. After heating the supernatant at 353 K, the protein was purified using anion-exchange chromatography. The purity and homogeneity were tested by SDS–PAGE. The protein was concentrated to 15 mg ml−1 in a Centricon YM-3 concentrator and stored at 277 K in a solution containing 20 mM Tris–HCl pH 7.8.
The selenomethionine (SeMet) derivative of SsEF-1β was prepared by growing the E. coli strain expressing the enzyme in minimal media containing 1 mg l−1 vitamins (riboflavin, niacinamide, pyridoxine monohydrochloride and thiamine), 0.4% glucose, 2 mM MgSO4, 0.1 mM CaCl2, 50 mg l−1 of the amino acids Phe, Thr, Lys, Ile, Leu and Val and 60 mg l−1 seleno-l-methionine. 5 mM DTT was added to all solutions used for purification of the SeMet derivative.
Crystallization experiments were performed at 293 K using either the microbatch-under-oil or hanging-drop vapour-diffusion methods. Preliminary crystallization trials were carried out using commercially available sparse-matrix screens (Crystal Screen and Wizard kits I and II from Hampton Research and deCode Genetics, respectively). Crystals suitable for X-ray diffraction were obtained by tuning the protein and precipitant concentrations.
A similar approach was used to grow crystals of the SeMet derivative. In this case, to prevent oxidation of the SeMet derivative, 5 mM DTT was added to the crystallization medium.
2.2. Data collection and processing
Preliminary diffraction data were collected in-house at 298 K using a MacScience DIP 2030b imaging plate equipped with a Nonius FR591 generator producing Cu Kα radiation of wavelength 1.5418 Å. Higher resolution data were collected at beamline BW7B of the DESY synchrotron (Hamburg, Germany) at 100 K. Crystals were flash-cooled after the addition of 20%(v/v) ethylene glycol to the crystallization buffer. Data processing was performed using the program DENZO (Otwinowski & Minor, 1997 ▶). The data sets were scaled and merged using the program SCALEPACK (Otwinowski & Minor, 1997 ▶).
Multiwavelength anomalous diffraction (MAD) data were collected at beamline ID29 of the ESRF synchrotron (Grenoble, France). Three different data sets were collected from a single crystal using wavelengths determined from the selenium-absorption spectrum.
2.3. Structure determination
Solution of the SsEF-1β structure using MAD methods is in progress. The program SOLVE (Terwilliger & Berendzen, 1999 ▶) was used to identify and localize the selenium sites present in the asymmetric unit and to derive the experimental phases. Phases were improved by density modification using the program DM (Cowtan & Main, 1998 ▶). Model building using both automatic and manual approaches is in progress.
3. Results and discussion
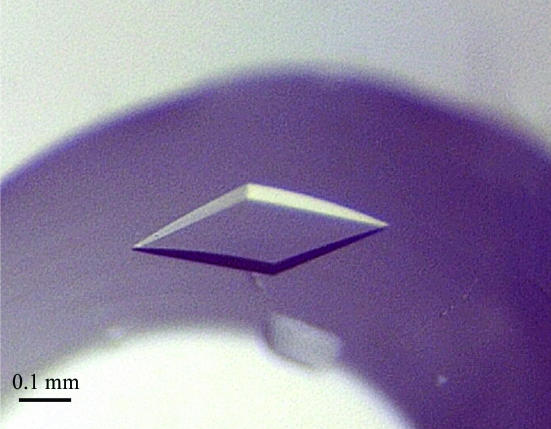
The initial screenings using commercially available solutions revealed several promising conditions for crystallization of SsEF-1β. All favourable conditions were characterized by the presence of polyethylene glycol as precipitating agent. The quality of the crystals was improved by fine-tuning the concentration of the protein and of the precipitants. SsEF-1β crystals (Fig. 1 ▶) suitable for X-ray diffraction data collection (0.3 × 0.2 × 0.1 mm) were obtained using 5–10 mg ml−1 protein solution and 9–10%(w/v) PEG 3000 as precipitant in 50 mM sodium acetate pH 4.5 buffer. Notably, crystals with a similar morphology were obtained using other rather different conditions. In particular, they were obtained in the presence of (i) 15%(w/v) PEG 8000 and 0.1 M (NH4)2SO4, (ii) 10–15%(w/v) PEG 8000, 100 mM lithium sulfate and 50 mM sodium acetate pH 4.5 and (iii) 15–20%(w/v) PEG 1000, 100 mM lithium sulfate and 50 mM phosphate–citrate buffer pH 4.2.
Figure 1.
Image of a typical SsEF-1β crystal grown using PEG 3000 as a precipitating agent (see text for details).
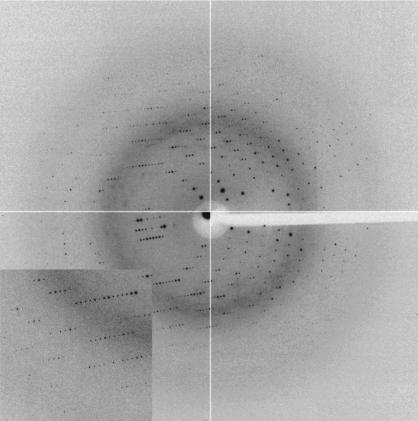
All these crystals proved to be isomorphous, with unit-cell parameters a = 106.46, b = 54.87, c = 44.03 Å (space group P21212). Using DESY synchrotron radiation, crystals of SsEF-1β diffracted to 1.97 Å (Fig. 2 ▶ and Table 1 ▶). Matthews coefficient calculations (Matthews, 1968 ▶) suggested the presence of either two (V M = 3.2 Å3 Da−1, with 62% solvent content) or three (V M = 2.1 Å3 Da−1, with 41% solvent content) SsEF-1β molecules per asymmetric unit. However, the self-rotation function did not exhibit any significant peak apart from that located at the origin.
Figure 2.
Diffraction pattern of a SsEF-1β crystal (SeMet derivative). Diffraction data are detectable to 1.8 Å resolution.
Table 1. Data-collection statistics.
Values given in parentheses refer to the highest resolution shell.
| SeMet derivative | ||||
|---|---|---|---|---|
| Crystal | Native | Peak | Edge | Remote |
| Beamline | BW7B, DESY | ID29, ESRF | ||
| Space group | P21212 | P21212 | ||
| Unit-cell parameters (Å) | ||||
| a | 106.46 | 105.72 | ||
| b | 54.87 | 54.69 | ||
| c | 44.03 | 43.42 | ||
| Resolution (Å) | 30–1.97 (2.04–1.97) | 30–1.83 (1.86–1.83) | 30–1.83 (1.86–1.83) | 30–1.83 (1.86–1.83) |
| Wavelength (Å) | 0.850 | 0.9792 | 0.9794 | 0.9756 |
| Mean redundancy | 3.6 | 3.8 | 3.8 | 3.8 |
| Completeness (%) | 90.4 (79.5) | 99.4 (97.1) | 99.4 (96.5) | 99.4 (97.1) |
| Unique reflections | 17141 | 42642 | 42600 | 42703 |
| Rmerge† (%) | 8.9 (37.3) | 5.7 (29.8) | 5.9 (35.4) | 5.9 (35.9) |
| Mean I/σ(I) | 12.9 (2.3) | 19.4 (3.0) | 18.9 (2.4) | 18.9 (2.3) |
R
merge = 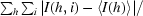
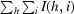 , where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the mean value of the intensity of reflection h. For the SeMet derivative the R
merge value has been calculated considering the Bijovet pairs individually.
, where I(h, i) is the intensity of the ith measurement of reflection h and 〈I(h)〉 is the mean value of the intensity of reflection h. For the SeMet derivative the R
merge value has been calculated considering the Bijovet pairs individually.
Several attempts were made to solve the structure by molecular replacement (MR) using S. cerevisiae and M. thermoautotrophicum EF-1β structures as starting models. However, all these attempts were unsuccessful. The failure of MR trials using the structure of ScEF-1β extracted from the ScEF-1α–EF-1β complex (PDB code 1f60; Andersen et al., 2000 ▶) can be ascribed to the very low sequence identity (<20%) and to the fact that the ScEF-1β structure may be affected by the formation of the complex with ScEF-1α. Moreover, taking into account that MR often fails when NMR models are used (Chen, 2001 ▶), it is not surprising that the use of MtEF-1β (PDB code 1gh8; Kozlov et al., 2000 ▶) did not yield correct solutions, despite the sequence identity shown by SsEF-1β and MtEF-1β (36%). In both cases the presence of two SsEF-1β molecules in the asymmetric unit of the crystal was a further difficulty for the MR approach.
Since MR proved to be unsuccessful, MAD experiments were carried out to obtain experimental phases. Therefore, an SeMet derivative of the protein was prepared. It is worth mentioning that, excluding the N-terminal methionine, a sole methionine is present in SsEF-1β sequence, which consists of 90 residues. Crystals of SeMet SsEF-1β were obtained from solutions containing 10–12%(w/v) PEG 8000, 50 mM lithium sulfate and 50 mM sodium acetate pH 4.5. In order to determine the peak and the inflection wavelengths, a fluorescence scan was recorded on a single SeMet-labelled SsEF-1β crystal. Using data sets collected at wavelengths optimized for selenomethionine, the program SOLVE (Terwilliger & Berendzen, 1999 ▶) identified two selenium sites in the asymmetric unit of the protein. This finding supports the hypothesis that two SsEF-1β molecules are present in the asymmetric unit. The program SOLVE also provided a set of initial phases. These phases were improved using the methods implemented in the program DM (Cowtan & Main, 1998 ▶). Using these improved phases, the program ARP/wARP (Perrakis et al., 1999 ▶) was able to automatically trace nearly 60% of the residues present in the asymmetric unit. Manual model-building sessions aimed at defining the complete SsEF-1β structure are in progress.
Acknowledgments
This work was financially supported by PRIN, MIUR 2003. We acknowledge the staff of the Macromolecular Crystallography Group at ESRF (beamline ID29, Grenoble, France) and the DESY staff (beamline BW7B, Hamburg, Germany) for providing the synchrotron-radiation facilities and for valuable assistance during data collection. Giosuè Sorrentino and Maurizio Amendola are acknowledged for their technical assistance.
References
- Andersen, G. R., Nissen, P. & Nyborg, J. (2003). Trends Biochem. Sci.28, 434–441. [DOI] [PubMed] [Google Scholar]
- Andersen, G. R., Pedersen, L., Valente, L., Chatterjee, I., Kinzy, T. G., Kjeldgaard, M. & Nyborg, J. (2000). Mol. Cell, 6, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Andersen, G. R., Valente, L., Pedersen, L., Kinzy, T. G. & Nyborg, J. (2001). Nature Struct. Biol.8, 531–534. [DOI] [PubMed] [Google Scholar]
- Arcari, P., Raimo, G., Ianniciello, G., Gallo, M. & Bocchini, V. (1995). Biochim. Biophys. Acta, 1263, 86–88. [DOI] [PubMed] [Google Scholar]
- Chen, Y. W. (2001). Acta Cryst. D57, 1457–1461. [DOI] [PubMed] [Google Scholar]
- Cherfils, J. & Chardin, P. (1999). Trends Biochem. Sci.24, 306–311. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. & Main, P. (1998). Acta Cryst. D54, 487–493. [DOI] [PubMed] [Google Scholar]
- Ianniciello, G., Masullo, M., Raimo, G., Arcari, P. & Bocchini, V. (1998). Protein Expr. Purif.12, 1–6. [DOI] [PubMed] [Google Scholar]
- Kozlov, G., Ekiel, I., Beglova, N., Yee, A., Dharamsi, A., Engel, A., Siddiqui, N., Nong, A. & Gehring, K. (2000). J. Biomol. NMR, 17, 187–194. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326.. [DOI] [PubMed]
- Perez, J. M., Siegal, G., Kriek, J., Hard, K., Dijk, J., Canters, G. W. & Moller, W. (1999). Structure Fold. Des.7, 217–226. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed] [Google Scholar]
- Raimo, G., Masullo, M. & Bocchini, V. (1999). FEBS Lett.451, 109–112. [DOI] [PubMed] [Google Scholar]
- Raimo, G., Masullo, M., Savino, G., Scarano, G., Ianniciello, G., Parente, A. & Bocchini, V. (1996). Biochim. Biophys. Acta, 1293, 106–112. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. (2002). Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]