The crystallization and preliminary X-ray diffraction analysis of the l-fuculose-1-phosphate aldolase (FucA) from T. thermophilus HB8. Native diffraction data set was collected to a resolution of 1.9 Å.
Keywords: fuculose phosphate aldolase, class II aldolases, Thermus thermophilus HB8
Abstract
Fuculose phosphate aldolase catalyzes the reversible cleavage of l-fuculose-1-phosphate to dihydroxyacetone phosphate and l-lactaldehyde. The protein from Thermus thermophilus HB8 is a biological tetramer with a subunit molecular weight of 21 591 Da. Purified FucA has been crystallized using sitting-drop vapour-diffusion and microbatch techniques at 293 K. The crystals belong to space group P4, with unit-cell parameters a = b = 100.94, c = 45.87 Å. The presence of a dimer of the enzyme in the asymmetric unit was estimated to give a Matthews coefficient (V M) of 2.7 Å3 Da−1 and a solvent content of 54.2%(v/v). Three-wavelength diffraction MAD data were collected to 2.3 Å from zinc-containing crystals. Native diffraction data to 1.9 Å resolution have been collected using synchrotron radiation at SPring-8.
1. Introduction
Aldolases have been divided into two groups of enzymes on the basis of their method of enzyme catalysis (Rutter, 1964 ▶). Class I aldolases are found predominantly in animals, higher plants and green algae, while class II aldolases are found in bacteria, yeasts and fungi. Class I aldolases have an essential lysine residue that forms a protonated Schiff base with the substrate carbonyl carbon to stabilize the intermediate (Horecker et al., 1972 ▶; Littlechild & Watson, 1993 ▶), whereas the class II aldolases are metal-containing enzymes in which the substrate is coordinated to a divalent metal cation such as zinc or magnesium (Rutter, 1964 ▶; Morse & Horecker, 1968 ▶). The potential of aldolases is widely recognized in synthetic chemistry (Wong et al., 1995 ▶; Whitesides & Wong, 1985 ▶), in particular the class II aldolases, which are more stable. Aldolases useful for organic synthesis can be classified into four groups based on the accepted donor substrate, namely (1) dihydroxyacetone phosphate as the donor to produce 2-keto-3,4-dihydroxy adducts, (2) pyruvate or phosphoenol pyruvate as the donor to form 3-deoxy-2-keto acids, (3) acetaldehyde as the donor to form 3-hydroxy aldehydes and (4) glycine as the donor to produce β-hydroxy-α-amino acids. These aldolases are the targets of inhibitors that may have antibacterial properties. Structures of class I aldolases have been previously determined from rabbit muscle (Sygusch et al., 1987 ▶; Blom & Sygusch, 1997 ▶), human muscle (Gamblin et al., 1991 ▶; Dalby et al., 1999 ▶), Drosophila melanogaster (Hester et al., 1991 ▶) and Plasmodium falciparum (Kim et al., 1998 ▶). Other aldolase structures have been determined from Pseudomonas putida and of N-acetylneuraminate aldolase from Escherichia coli (Izard et al., 1994 ▶; Lawrence et al., 1997 ▶), 7,8-dihydroneopterin aldolase from Staphylococcus aureus (Hennig et al., 1998 ▶), l-rhamnulose-1-phosphate aldolase (Kroemer & Schulz, 2002 ▶), fructose-1,6-bisphosphate aldolase from the hyperthermophile Thermoproteus tenax (Lorentzen et al., 2003 ▶) and 2-deoxyribose-5-phosphate aldolase from Thermus thermophilus HB8, E. coli and Aeropyrum pernix (Lokanath et al., 2004 ▶; Heine et al., 2001 ▶; Sakuraba et al., 2003 ▶). Class II aldolases contain a divalent metal ion (usually zinc). The known structures of this class include l-fuculose-1-phosphate aldolase from E. coli (Dreyer & Schulz, 1993 ▶, 1996a ▶,b ▶) and fructose-1,6-bisphosphate aldolase from E. coli (Blom et al., 1996 ▶; Cooper et al., 1996 ▶; Hall et al., 1999 ▶). l-Fuculose-1-phosphate from T. thermophilus HB8 (FucA) is a class II aldolase that catalyzes the reversible cleavage of l-fuculose-1-phosphate (Fuc1P), leading to dihydroxyacetone phosphate (DHAP) and l-lactaldehyde, which is a crucial step in the bacterial l-fucose metabolism. This reaction constitues one of the most important methods for forming carbon–carbon bonds (Ghalambor & Heath, 1962 ▶) in synthetic organic chemistry. We report here the purification, crystallization and preliminary X-ray analysis of fuculose-l-phosphate aldolase (FucA) from T. thermophilus HB8.
2. Material and methods
2.1. Protein expression and purification
The gene was amplified by the polymerase chain reaction (PCR) using T. thermophilus HB8 genomic DNA as a template. The recombinant plasmid was constructed using the super-rare-cutter system (Kanagawa et al., manuscript in preparation). E. coli BL21-CodonPlus (DE3)-RIL cells were transformed with the recombinant plasmid and grown at 310 K in LB medium containing 50 µg ml−1 ampicillin for 20 h. The cells were harvested by centrifugation at 6500 rev min−1 for 5 min at 277 K and were suspended in 20 mM Tris–HCl pH 8.0 (buffer A) containing 0.5 M NaCl and 5 mM 2-mercaptoethanol and disrupted by sonication. The cell lysate was clarified by centrifugation (15 000 rev min−1, 30 min) and the supernatant was heated at 363 K for 11.5 min. After heat treatment, denaturated proteins were removed by centrifugation (15 000 rev min−1, 30 min) and the supernatant solution was used as the crude extract for purification. The crude extract was desalted using HiPrep 26/10 desalting column (Amersham Biosciences) and applied onto a Super Q Toyopearl 650M column (Tosoh) equilibrated with buffer A. The protein was eluted with a linear gradient of 0–0.3 M NaCl in buffer A. The protein was desalted using a HiPrep 26/10 desalting column with buffer A and subjected to a Resource Q column (Amersham Biosciences) equilibrated with buffer A. The protein was eluted with a linear gradient of 0–0.3 M NaCl in buffer A. The fractions containing proteins were desalted using a HiPrep 26/10 desalting column with 10 mM sodium phosphate pH 7.0 and applied onto a Bio-Scale CHT-20-I column (Bio-Rad) equilibrated with the same buffer. The protein was eluted with a linear gradient of 10–300 mM sodium phosphate pH 7.0. The fractions containing protein were pooled, concentrated by ultrafiltration (Vivaspin, 5 kDa cutoff) and loaded onto a HiLoad 16/60 Superdex 200 prep-grade column (Amersham Biosciences) equilibrated with buffer A containing 0.2 M NaCl. The purified protein was homogeneous on native PAGE.
2.2. Crystallization
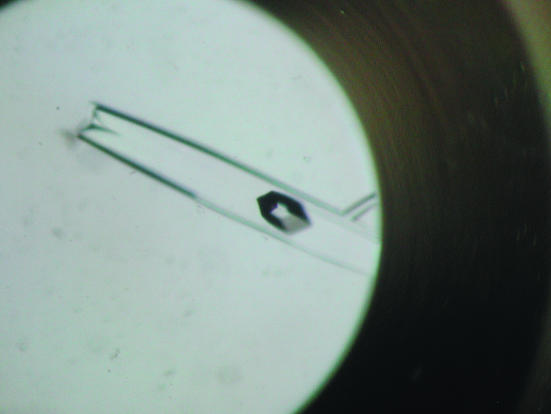
Crystallization was performed by the sitting-drop vapour-diffusion method at 293 K using Linbro multiwell plates. Each drop consisting of 1 µl 20 mg ml−1 protein solution in 0.1 M Tris–HCl buffer pH 7.9 and 1 µl reservoir solution was allowed to equilibrate against 100 µl reservoir solution. Preliminary screening was performed using Hampton Research Crystal Screens I and II (Jancarik & Kim, 1991 ▶). Small crystals appeared from a condition with reservoir solution consisting of 1.5 M lithium sulfate monohydrate in 0.1 M HEPES pH 7.5. Slightly larger crystals were obtained in the range 1.4–1.5 M lithium sulfate monohydrate, 0.1 M Na HEPES pH 7.0–7.5, followed by refinement of this condition through variation of protein concentration, pH and drop volume. To improve the crystal size and quality, further optimization of the conditions was carried out using the microbatch (diffusion through paraffin oil) method with Nunc HLA plates (Nalge Nunc International) at 293 K. Crystals suitable for X-ray data collection appeared (Fig. 1 ▶) within 10–15 d and reached final dimensions of 0.6 × 0.3 × 0.1 mm.
Figure 1.
A crystal (two crystals are inside the well) of l-fuculose-1-phosphate aldolase from T. thermophilus HB8 (FucA). The largest crystal has approximate dimensions of 0.6 × 0.3 × 0.1 mm.
2.3. Data collection and processing
Diffraction data were collected using a Jupiter 210cs CCD detector at the BL26B1 beamline, SPring-8, Japan. The crystals were flash-frozen in a nitrogen-gas stream at 100 K directly from a drop containing 30%(w/v) glycerol as a cryoprotectant and were maintained at 100 K during data collection. For the Zn-containing crystal, MAD (multiple anomalous diffraction) data sets were collected corresponding to the maximum 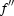 (peak), the minimum
(peak), the minimum 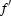 (edge) and a reference wavelength (remote), chosen on the basis of the absorption spectrum of the metal. Based on the absorption spectrum, two energy levels were chosen for data collection, both of which were near the absorption K edge of the Zn atom: 9.669 and 9.666 keV. The third energy level was set to 9.301 keV as a remote point. Native diffraction data were collected to 1.9 Å. The diffraction data were processed with the HKL2000 package (Otwinowski & Minor, 1997 ▶). The crystallographic data and the MAD data statistics for zinc-containing crystals are summarized in Table 1.
(edge) and a reference wavelength (remote), chosen on the basis of the absorption spectrum of the metal. Based on the absorption spectrum, two energy levels were chosen for data collection, both of which were near the absorption K edge of the Zn atom: 9.669 and 9.666 keV. The third energy level was set to 9.301 keV as a remote point. Native diffraction data were collected to 1.9 Å. The diffraction data were processed with the HKL2000 package (Otwinowski & Minor, 1997 ▶). The crystallographic data and the MAD data statistics for zinc-containing crystals are summarized in Table 1.
2.4. Dynamic light-scattering studies
A dynamic light-scattering experiment was performed using a DynaPro MS/X instrument from Protein Solutions (Lakewood, New Jersey, USA). The measurements were made at 291 K on the purified protein at 0.5–1.0 mg ml−1 in buffer solution containing 20 mM Tris–HCl and 200 mM sodium chloride.
3. Discussion
The crystals belonged to space group P4, with unit-cell parameters a = b = 100.94, c = 45.87 Å. A total of 245 580 measured reflections in the resolution range 30–1.9 Å were merged into 36 580 unique reflections with an R merge of 6.6%. Details of data collection and processing are given in Table 1 ▶. The value of the Matthews coefficient (Matthews, 1968 ▶) is 2.7 Å3 Da−1 and the solvent content is 54.2%(v/v) assuming the presence of a dimer in the asymmetric unit. Dynamic light-scattering experiments showed the presence of a tetramer in solution, suggesting biological significance of the tetramer. The present enzyme shows 33% sequence identity with l-fuculose-1-phosphate aldolase from E. coli (PDB code 1dzw; Dreyer & Schulz, 1993 ▶). The molecular-replacement method using l-fuculose-1-phosphate aldolase as a search model, which was attempted using AMoRe (Navaza, 1994 ▶) and PHASER (Read, 2001 ▶; Storoni et al., 2004 ▶), did not yield a clear solution. Class II aldolases contain a divalent metal ion (usually zinc) in the active site. On the basis of this, we measured the X-ray fluorescence scans of the divalent metal atoms Fe2+, Ni2+, Co2+ and Zn2+. X-ray fluorescence scans of the crystals had a strong absorption edge corresponding to Zn. The diffraction data recorded at wavelengths near the Zn X-ray absorption edge were used in MAD phasing (Hendrickson et al., 1990 ▶). The peak data set was used for the analysis of Bijvoet and difference Patterson maps did not clearly show the position of the anomalous scatter. The average value of the anomalous signal-to-noise ratio in the high-energy data set is 1.69 in the resolution range 30–2.3 Å and is similar to that in the edge and remote data sets. Therefore, reliable initial phases could not be obtained from the Zn MAD data sets. A selenomethionyl derivative of the protein has now been obtained under the same conditions as used for the native crystals and appears to have similar diffraction properties. Further attempts to determine the structure using Zn data as well as SeMet data are in progress.
Table 1. Experimental conditions and data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | Native | Zn peak | Edge | Remote |
|---|---|---|---|---|
| X-ray source | BL26B1, SPring-8 | BL26B1, SPring-8 | ||
| Wavelength (Å) | 1.0000 | 1.28230 | 1.28275 | 1.3330 |
| Detector | Jupiter 210cs CCD | Jupiter 210cs CCD | ||
| Temperature (K) | 100 | 100 | ||
| Crystal-to-detector distance (mm) | 180 | 150 | ||
| Space group | P4 | P4 | ||
| Unit-cell parameters (Å) | a = b = 100.944, c = 45.870 | a = b = 100.860, c = 45.959 | ||
| Resolution range (Å) | 30.0–1.90 (1.97–1.90) | 50–2.30 (2.38–2.30) | ||
| Total reflections | 245580 | 105929 | 100444 | 102657 |
| Unique reflections | 36580 | 20641 | 20603 | 20593 |
| Completeness (%) | 99.3 (99.2) | 98.8 (99.2) | 98.8 (99.3) | 98.7 (99.4) |
| Rsym† | 6.6 (24.6) | 10.8 (26.0) | 10.6 (25.3) | 11.6 (28.5) |
| Average 〈I/σ(I)〉 | 18.4 (3.8) | 17.9 (5.8) | 17.1 (5.2) | 16.8 (5.0) |
| Redundancy | 6.7 | 5.1 | 4.9 | 5.0 |
| Anomalous signal (asn‡) | 1.69 | |||
| VM (Å3 Da−1) | 2.7 | |||
| Z | 2 | |||
| Solvent content (%) | 54.2 | |||
R
sym = 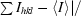
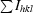 .
.
Average anomalous signal-to-noise ratio.
Acknowledgments
The authors would like to thank the staff of RIKEN Genomic Science Centre, Yokohama for providing the plasmid. We also thank M. Yamamoto and his staff for assistance during data collection at beamline BL26B1 of SPring-8. This work was supported by a National Project on Protein Structural and Functional Analysis funded by MEXT of Japan (Project PH0500/HTPF00294).
References
- Blom, N. & Sygusch, J. (1997). Nature Struct. Biol.4, 36–39. [DOI] [PubMed] [Google Scholar]
- Blom, N. S., Tétreault, S., Coulombe, R. & Sygusch, J. (1996). Nature Struct. Biol.3, 856–862. [DOI] [PubMed] [Google Scholar]
- Cooper, S. J., Leonard, G. A., McSweeney, S. M., Thompson, A. W., Naismith, J. H., Qamar, S., Plater, A., Berry, A. & Hunter, W. N. (1996). Structure, 4, 1303–1315. [DOI] [PubMed] [Google Scholar]
- Dalby, A., Dauter, Z. & Littlechild, J. A. (1999). Protein Sci.8, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer, M. K. & Schulz, G. E. (1993). J. Mol. Biol.231, 549–553. [DOI] [PubMed] [Google Scholar]
- Dreyer, M. K. & Schulz, G. E. (1996a). Acta Cryst. D52, 1082–109. [DOI] [PubMed] [Google Scholar]
- Dreyer, M. K. & Schulz, G. E. (1996b). J. Mol. Biol.259, 458–466. [DOI] [PubMed] [Google Scholar]
- Gamblin, S. J., Davies, G. J., Grimes, J. M., Jackson, R. M., Littlechild, J. A. & Watson, H. C. (1991). J. Mol. Biol.219, 573–576. [DOI] [PubMed] [Google Scholar]
- Ghalambor, M. A. & Heath, E. C. (1962). J. Biol. Chem.237, 2427–2433. [PubMed] [Google Scholar]
- Hall, D. R., Leonard, G. A., Reed, C. D., Watt, C. I., Berry, A. & Hunter, W. N. (1999). J. Mol. Biol.287, 383–394. [DOI] [PubMed] [Google Scholar]
- Heine, A., DeSantis, G., Luz, J. G., Mitchell, M., Wong, C.-H. & Wilson, I. A. (2001). Science, 294, 369–374. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990). EMBO J.9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, M., D’Arcy, A., Hampele, I. C., Page, M. G. P., Oefner, C. & Dale, G. E. (1998). Nature Struct. Biol.5, 357–362. [DOI] [PubMed] [Google Scholar]
- Hester, G., Brenner-Holzach, O., Rossi, F. A., Struck-Donatz, M., Winterhalter, K. H., Smit, J. D. G. & Piontek, K. (1991). FEBS Lett.292, 237–242. [DOI] [PubMed] [Google Scholar]
- Horecker, B. L., Tsolas, O. & Lai, C. Y. (1972). The Enzymes, edited by P. D. Boyer, 3rd ed., Vol. 7, pp. 213–258. New York: Academic Press.
- Izard, T., Lawrence, M. C., Malby, R., Lilley, G. G. & Colman, P. M. (1994). Structure, 2, 361–369. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kim, H., Certa, U., Dobeli, H., Jakob, P. & Hol, W. G. J. (1998). Biochemistry, 37, 4388–4396. [DOI] [PubMed] [Google Scholar]
- Kroemer, M. & Schulz, G. E. (2002). Acta Cryst. D58, 824–832. [DOI] [PubMed] [Google Scholar]
- Lawrence, M. C., Barbosa, J. A. R. G., Smith, B. J., Hall, N. E., Pilling, P. A., Ooi, H. C. & Marcuccio, S. M. (1997). J. Mol. Biol.266, 381–399. [DOI] [PubMed] [Google Scholar]
- Littlechild, J. A. & Watson, H. C. (1993). Trends Biochem. Sci.18, 36–39. [DOI] [PubMed] [Google Scholar]
- Lokanath, N. K., Shiromizu, I., Ohshima, N., Nodake, Y., Sugahara, M., Yokoyama, S., Kuramitsu, S., Miyano, M. & Kunishima, N. (2004). Acta Cryst. D60, 1816–1823. [DOI] [PubMed] [Google Scholar]
- Lorentzen, E., Pohl, E., Zwart, P., Stark, A., Russell, R. B., Knura, T., Hensel, R. & Siebers, B. (2003). J. Biol. Chem.278, 47253–47260. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Morse, D. E. & Horecker, B. L. (1968). Adv. Enzymol. Relat. Areas Mol. Biol.31, 125–181. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Rutter, W. J. (1964). Fed. Proc.23, 1248–1257. [PubMed] [Google Scholar]
- Sakuraba, H., Tsuge, H., Shimoya, I., Kawakami, R., Goda, S., Kawarabayasi, Y., Katunuma, N., Ago, H., Miyano, M. & Ohshima, T. (2003). J. Biol. Chem.278, 10799–10806. [DOI] [PubMed] [Google Scholar]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed] [Google Scholar]
- Sygusch, J., Beaudry, D. & Allaire, M. (1987). Proc. Natl Acad. Sci. USA, 84, 7846–7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides, G. M. & Wong, C. H. (1985). Angew. Chem. Int. Ed. Engl.24, 617–638.
- Wong, C. H., Halcomb, R. H., Ichikawa, Y. & Kajimoto, T. (1995). Angew. Chem. Int. Ed. Engl.34, 412–432.