The first crystallographic study of an isolated WW domain is reported. Single crystals of the WW4 domain of the Nedd4-2 ubiquitin–protein ligase contain a high solvent content of 74% and diffract X-rays to 2.5 Å resolution.
Keywords: Nedd4-2 ubiquitin–protein ligase, WW4 domain
Abstract
Ubiquitin-mediated protein modification via covalent attachment of ubiquitin has emerged as one of the most common regulatory processes in all eukaryotes. Nedd4-2, closely related to neuronal precursor cell-expressed developmentally down-regulated 4 (Nedd4), is a multimodular ubiquitin–protein ligase comprised of four WW domains and a Hect domain. The WW domains recognize the proline-rich motifs on the multi-subunit amiloride-sensitive epithelial sodium channel (ENaC). To gain insights into the binding of the WW domain to proline-rich peptides, a protein fragment (78 amino acids) containing the fourth WW domain (WW4) of the Nedd4-2 protein was purified and crystallized and X-ray diffraction data were collected. A data set was obtained to 2.5 Å resolution from a cryocooled single crystal at a synchrotron source. The crystals belong to the tetragonal space group P41212 (or P43212), with unit-cell parameters a = b = 113.43, c = 103.21 Å. Analysis of the self-rotation function suggests the presence of four WW4 molecules in the asymmetric unit, with a high unit-cell solvent content of 74%.
1. Introduction
Ubiquitin–protein ligases play an important role in signalling pathways and disease processes ranging from developmental abnormalities and autoimmunity to neurodegenerative diseases and cancer (Hershko & Ciechanover, 1998 ▶; Weissman, 2001 ▶). Ubiquitylation is a multistep process involving the successive action of three types of enzymes: E1, E2 and E3. Ubiquitin–protein ligase (E3) catalyzes the transfer of ubiquitin from the E2 enzyme to the ∊-amino group of a lysine residue on the substrate. E3, either alone or in combination with its bound E2, determines the exquisite sensitivity of substrate recognition (Hershko & Ciechanover, 1998 ▶). The RING and Hect types of E3 constitute the two major families of ubiquitin–protein ligases (Weissman, 2001 ▶). The amino-terminal sequences of Hect E3s are not conserved and contain the primary determinants for specific substrate recognition. Neuronal precursor cell-expressed developmentally down-regulated 4 protein (Nedd4) is a ubiquitin–protein ligase (E3) and is a prototypic molecule of an emerging family of Nedd4-related proteins (Kumar et al., 1997 ▶; Harvey & Kumar, 1999 ▶).
The Nedd4 family of proteins is involved in diverse cellular functions, such as regulation of membrane channels and permeases, the cell cycle and transcription (Harvey & Kumar, 1999 ▶; Ingham et al., 2004 ▶). Nedd4 was originally identified as a developmentally regulated mouse gene highly expressed in the early embryonic central nervous system (Kumar et al., 1992 ▶). Human Nedd4 has a modular structure consisting of an amino-terminal Ca2+/lipid-binding domain followed by four WW domains (WW1–WW4) and a carboxy-terminal ubiquitin–protein ligase domain (Hect E3) (Harvey & Kumar, 1999 ▶; Ingham et al., 2004 ▶). Nedd4 is known to target the epithelial sodium channel (ENaC) and voltage-gated sodium channels (Navs) for degradation (Thomas & Itani, 2004 ▶; Fotia et al., 2004 ▶). ENaC plays a major role in regulating blood pressure, and mutations in ENaC that disrupt the interaction with Nedd4 cause a human hypertensive disorder, Liddle’s syndrome (Thomas & Itani, 2004 ▶). Recent studies of Nedd4-2 protein, which is closely related to Nedd4, suggest that Nedd4-2 is the physiological regulator of ENaC and Navs (Harvey et al., 2001 ▶; Kamynina et al., 2001 ▶; Fotia et al., 2003 ▶, 2004 ▶). Unlike Nedd4, all Nedd4-2 proteins from various species including human contain four WW domains followed by a Hect domain (Harvey et al., 2001 ▶; Kamynina et al., 2001 ▶). The interaction of WW domains with the PY motifs of ENaC subunits is proposed to mediate ENaC ubiquitination by the Hect domain of Nedd4 proteins.
WW domains derive their name from the presence of two highly conserved tryptophan residues in a sequence of ∼40 amino acids. These domains are protein–protein interaction motifs that bind to proline-rich sequences called PY motifs (Sudol & Hunter, 2000 ▶), of which the PPXY motif is the most common. Binding studies between the WW domains of Nedd4-2 protein and ENaC subunits containing PY motifs indicate the requirement of WW3 and WW4 for both binding to ENaC subunits and regulation of Na+ feedback control of ENaC in vivo (Fotia et al., 2003 ▶). Domains WW1 and WW2 of Nedd4-2 do not show appreciable affinity for ENaC subunits. An understanding of the precise atomic interactions between the WW domains and the target PY motifs is essential to enhance our knowledge of the principles of molecular recognition of proline-rich peptides. Towards this goal, we have initiated structural studies of the WW domain with and without bound peptides. Here, we describe purification, crystallization and X-ray data collection for the WW4 domain of Nedd4-2 protein. The protein fragment used in the present study contains 78 amino-acid residues with a molecular weight of approximately 8.6 kDa. This is the first reported crystallographic work for an isolated WW domain.
2. Methods and results
2.1. Preparation of WW4 domain of Nedd4-2
A WW4 domain-glutathione S-transferase (GST) construct (Fotia et al., 2003 ▶) was generated by PCR amplification of WW4 domain from mouse Nedd4-2 cDNA followed by cloning into the EcoRI site of pGEX-2TK (Amersham Biosciences). This cloned plasmid was transformed into Escherichia coli strain BL21. Overnight cultures of E. coli harbouring the GST-WW4 expression plasmid were diluted 1:50, grown for 2 h at 310 K, induced with 1 mM isopropyl β-d-thiogalactoside and grown for an additional 4 h. Bacterial cell pellets were resuspended in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.3) with 1 mM DTT and 1 mM PMSF, sonicated and centrifuged at 10 000g for 20 min. Glutathione-Sepharose (Amersham Biosciences) was incubated with the cleared lysate for 1 h at room temperature and the beads were then washed three times with PBS buffer. The fusion protein was eluted with glutathione buffer as described previously (Jolliffe et al., 2000 ▶). The GST-WW4 fusion protein was cleaved by incubation with bovine thrombin according to the manufacturer’s protocol to obtain the WW4 domain. The WW4 domain was then purified to homogeneity by gel-filtration chromatography on a HiLoad 16/60 Superdex 30 column (Amersham Biosciences) in PBS buffer with 1 mM DTT. This WW4 sample eluted as a single peak when analyzed by reverse-phase HPLC (C18) and migrated as a single band on SDS–PAGE. The protein was concentrated to 8 mM using Amicon stirred cells with Millipore ultrafiltration membrane (MWCO of 3 kDa) and stored in the same PBS buffer.
2.2. Crystallization of WW4 domain
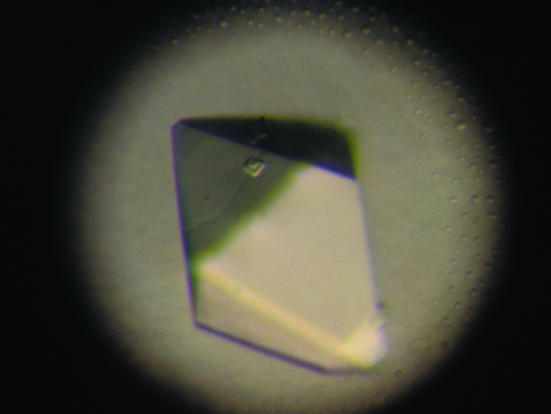
Initial crystallization trials were performed with commercially available Hampton solutions. Screening was carried out in 24-well trays using hanging drops containing 2 µl protein solution plus 2 µl reservoir solution set up for vapour diffusion against 500 µl reservoir solution at 277 and 293 K. Subsequently, custom-designed crystallization experiments were performed. Crystals of WW4 domain were obtained at room temperature in about two weeks. The crystallization droplet contained 8 mM protein, 50 mM sodium citrate pH 6.0, 1 mM EDTA, 1 mM DTT and 15%(w/v) PEG 10K. The reservoir contained 30%(w/v) PEG 10K and 100 mM sodium citrate buffer pH 6.0. The reproducibility of crystals was low and most crystallization trials resulted in overlapped crystals. Only a few crystallization experiments yielded small, isolated and fragile crystals. Repeated seeding trials resulted in larger crystals suitable for X-ray diffraction experiments. Prior to data collection, crystals were dipped in reservoir solution with 20% glycerol as cryoprotectant and flash-cooled in liquid nitrogen. Although of good appearance and large size, these crystals diffracted poorly to approximately 3.5 Å resolution at a synchrotron facility. Consequently, further work including crystallization trials with various additives and flash-cooling crystals using different cryoprotectants such as MPD, PEG 400 and d-glucose was undertaken with the aim of obtaining better diffraction. However, none of these trials improved the diffraction resolution. Several large crystals obtained by seeding were screened for diffraction, of which one crystal measuring 0.2 × 0.2 × 0.5 mm (Fig. 1 ▶) diffracted to 2.5 Å resolution.
Figure 1.
Photograph of a crystal of the WW4 domain of Nedd4-2 protein. A single crystal with dimensions 0.2 × 0.2 × 0.5 mm was used for data collection to 2.5 Å resolution.
2.3. X-ray diffraction
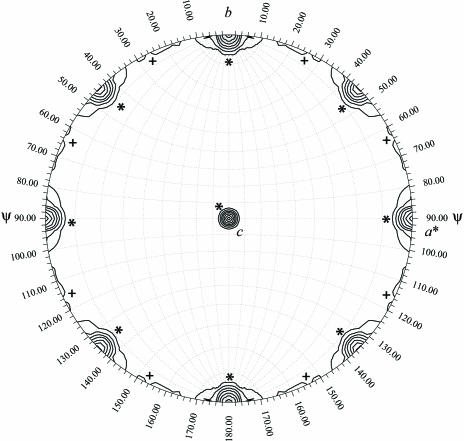
A crystal cooled using glycerol as a cryoprotectant was used to collect X-ray data at a synchrotron source using an ADSC Quantum 4 detector. The data were processed with DENZO and scaled using SCALEPACK (Otwinowski & Minor, 1997 ▶). The crystals belong to a tetragonal space group with unit-cell parameters a = b = 113.43, c = 103.21 Å. The volume of the unit cell is huge (Table 1 ▶) for the size of the molecular fragment (∼8.6 kDa), indicating a large number of molecules and/or a high solvent content. There is no evidence of oligomerization of this fragment in solution. In order to estimate the number of molecules in the asymmetric unit, we tried to measure the experimental density of the crystals using organic solvents such as ethanol and acetone, but the crystals degraded in the mixture of solvents. Analysis of the self-rotation function (Tong & Rossmann, 1997 ▶) using 10–3.5 Å resolution data and a radius of integration of 20 Å suggests the presence of four molecules in the asymmetric unit (Fig. 2 ▶) with a solvent content of 74% and a V M of 4.8 Å3 Da−1, a value well above the normal range observed for most proteins (Matthews, 1968 ▶). Some examples exhibiting very high solvent content include colicin Ia (78%; Wiener et al., 1997 ▶) and archaeal virus resolvase SIRV2 (77%; Ennifar et al., 2005 ▶). Although it is not always correct to correlate high solvent content with poor diffraction, in the present case we presume that the generally poor diffraction quality (<4.0 Å resolution) of even large crystals (∼0.2 × 0.2 × 0.5 mm) may in part be a consequence of the very high solvent content. Further optimization of cryoprotection, dehydration and crystallization protocols may help in improving the diffraction properties of these crystals.
Table 1. Summary of X-ray diffraction data for the WW4 domain of Nedd4-2 protein.
Values in parentheses are for the last resolution shell.
| Temperature (K) | 100 |
| X-ray source | Synchrotron/CHESS F1 |
| Wavelength (Å) | 0.9124 |
| Resolution range (Å) | 20–2.5 (2.59–2.5) |
| Space group | P41212 (or P43212) |
| Unit-cell volume (Å3) | 1.33 × 106 |
| No. of measured reflections | 250845 |
| No. of unique reflections | 23340 |
| Completeness of data (%) | 97.2 (72.1) |
| Mosaicity (°) | 0.43 |
| Rsym† (%) | 9.3 (15.5) |
R
sym = 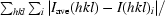
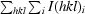 .
.
Figure 2.
Self-rotation function with κ = 180°. The rotation represented by a set of polar angles (ϕ, ψ and κ) was calculated with GLRF (Tong & Rossmann, 1997 ▶) using diffraction data between 10 and 3.5 Å resolution. Contour lines are drawn starting at 4 r.m.s. (root-mean-square) deviations above the mean in intervals of 1 r.m.s. deviation. Crystallographic peaks (between 6 and 11 r.m.s deviations) are marked with a star (*) and non-crystallographic peaks (∼5 r.m.s. deviations) with a plus sign (+).
Structure determination using molecular replacement based on a model corresponding to the WW segment from the crystal structure of Pin1–phosphopeptide complex (PDB code 1f8a; Verdecia et al., 2000 ▶) is under way. However, because the model for molecular replacement accounts for less than 50% of the size of the protein fragment containing the WW4 domain used here, the chances of a successful rotation and translation searches are slim. Therefore, we are also performing trials to obtain heavy-atom derivatives by treatment of crystals with sodium bromide solution as well as traditional heavy-atom soaking to determine experimental phases. Additionally, we are preparing selenomethionine-derivatized protein for MAD phasing. Furthermore, cocrystallization and soaking experiments to obtain crystals of a WW4–peptide complex are in progress. The WW4 domain of Nedd4-2 protein binds to α-, β- and γ-ENaC subunits with dissociation constants ranging from 0.77 to 2.1 µM. The PPXY motifs present in the α-ENaC (LALTAPPPAYATLGPS), β-ENaC (PIPGTPPPNYDSLRLQ) and γ-ENaC (PVPGTPPPRYNTLRLD) subunits have been used in binding studies (Fotia et al., 2003 ▶). In our crystallization trials, we have used these 16-mer peptides and their truncated versions, a rationale normally adopted in the preparation of protein–peptide complexes (Waksman et al., 1993 ▶).
Acknowledgments
This work was supported by grant No. IRG-91-022-09 from the American Cancer Society. NN thanks Dr Weiss for support and encouragement. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health through its National Center for Research Resources.
References
- Ennifar, E., Basquin, J., Birkenbihl, R. & Suck, D. (2005). Acta Cryst. F61, 507–509. [DOI] [PMC free article] [PubMed]
- Fotia, A. B., Dinudom, A., Shearwin, K. E., Koch, J. P., Korbmacher, C., Cook, D. I. & Kumar, S. (2003). FASEB J.17, 70–72. [DOI] [PubMed] [Google Scholar]
- Fotia, A. B., Ekberg, J., Adams, D. J., Cook, D. I., Poronnik, P. & Kumar, S. (2004). J. Biol. Chem.279, 28930–28935. [DOI] [PubMed] [Google Scholar]
- Harvey, K. F., Dinudom, A., Cook, D. I. & Kumar, S. (2001). J. Biol. Chem.276, 8597–8601. [DOI] [PubMed] [Google Scholar]
- Harvey, K. F. & Kumar, S. (1999). Trends Cell Biol.9, 166–169. [DOI] [PubMed] [Google Scholar]
- Hershko, A. & Ciechanover, A. (1998). Annu. Rev. Biochem.67, 425–479. [DOI] [PubMed] [Google Scholar]
- Ingham, R. J., Gish, G. & Pawson, T. (2004). Oncogene, 23, 1972–1984. [DOI] [PubMed] [Google Scholar]
- Jolliffe, C. N., Harvey, K. F., Haines, B. P., Parasivam, G. & Kumar, S. (2000). Biochem. J.351, 557–565. [PMC free article] [PubMed] [Google Scholar]
- Kamynina, E., Debonneville, C., Bens, M., Vandewalle, A. & Staub, O. (2001). FASEB J.15, 204–214. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Harvey, K. F., Kinoshita, M., Copeland, N. G., Noda, M. & Jenkins, N. A. (1997). Genomics, 40, 435–443. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tomooka, Y. & Noda, M. (1992). Biochem. Biophys. Res. Commun.185, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sudol, M. & Hunter, T. (2000). Cell, 103, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Thomas, C. P. & Itani, O. A. (2004). Curr. Opin. Nephrol. Hypertens.13, 541–548. [DOI] [PubMed] [Google Scholar]
- Tong, L. & Rossmann, M. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]
- Verdecia, M. A., Bowman, M. E., Lu, K. P., Hunter, T. & Noel, J. P. (2000). Nature Struct. Biol.7, 639–643. [DOI] [PubMed] [Google Scholar]
- Waksman, G., Shoelson, S. E., Pant, N., Cowburn, D. & Kuriyan, J. (1993). Cell, 72, 779–790. [DOI] [PubMed] [Google Scholar]
- Weissman, A. M. (2001). Nature Rev. Mol. Cell Biol.2, 169–178. [DOI] [PubMed]
- Wiener, M., Freymann, D., Ghosh, P. & Stroud, R. M. (1997). Nature (London), 385, 461–464. [DOI] [PubMed] [Google Scholar]