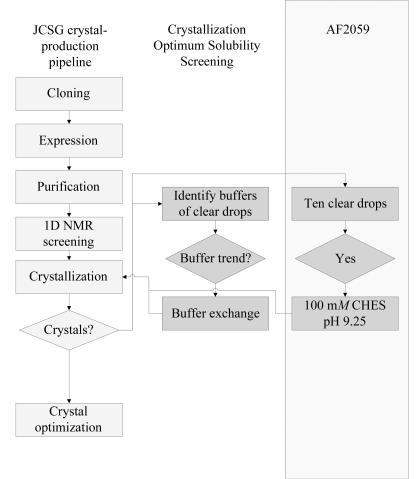
It is shown how protein crystallization results can be used to identify buffers that improve protein solubility and, in turn, crystallization success.
Keywords: crystallization, optimum solubility screen, buffer, high throughput, structural genomics
Abstract
An optimal solubility screen is described that uses the results of crystallization trials to identify buffers that improve protein solubility and, in turn, crystallization success. This screen is useful not only for standard crystallization experiments, but also can easily be implemented into any high-throughput structure-determination pipeline. As a proof of principle, the predicted novel-fold protein AF2059 from Archaeoglobus fulgidus, which was known to precipitate in most buffers and particularly during concentration experiments, was selected. Using the crystallization results of 192 independent crystallization trials, it was possible to identify a buffer containing 100 mM CHES pH 9.25 that significantly improves its solubility. After transferring AF2059 into this ‘optimum-solubility’ buffer, the protein was rescreened for crystal formation against these same 192 conditions. Instead of extensive precipitation, as observed initially, it was found that 24 separate conditions produced crystals and the exchange of AF2059 into CHES buffer significantly improved crystallization success. Fine-screen optimization of these conditions led to the production of a crystal suitable for high-resolution (2.2 Å) structure determination.
1. Introduction
Screening for suitable protein constructs and crystallization conditions remain the rate-limiting steps in protein structure determination owing to the extensive number of variables that can be systematically altered (McPherson, 1994 ▶). This is especially true for the high-throughput (HT) structural genomics (SG) community, where detailed optimization of expression, purification and crystallization conditions for individual proteins is more difficult to implement because of the use of parallel robotics for most steps of the protein structure-determination process (Lesley et al., 2002 ▶). One step to maximize the likelihood of obtaining diffraction-quality crystals is to identify the buffer in which the protein of interest forms a soluble monodisperse sample (D’Arcy, 1994 ▶; Wilson, 2003 ▶). Jancarik et al. (2004 ▶) have developed an elegant screening protocol, optimum solubility (OS) screening, to identify the optimal buffer for protein stability prior to crystallization trials. Briefly, proteins are screened against 24 distinct buffers and examined for lack of precipitate formation. The proteins are mixed with equal volumes of buffer and set up in drops identical to those typically used for crystallization trials. After overnight equilibration at room temperature, the drops that are clear are then diluted with an additional 15 µl of buffer and the resulting samples screened by dynamic light scattering (DLS) to determine the aggregation state of the protein. Those buffer(s) in which the protein is soluble and monodisperse are then selected as the optimal protein-equilibration buffer for subsequent crystallization trials.
We have developed an alternative optimal solubility screening method that does not require an additional screening step to be implemented into the structure-determination process. Instead, the results of initial crystallization trials are used to identify the optimum buffer for a given protein for crystallization (Fig. 1 ▶). Specifically, instead of screening 24 buffers, as described for the Jancarik optimum solubility screen, the results of the initial crystallization trials are analyzed, the clear drops (those without crystals/precipitate) are identified and the crystallization components of the clear drops evaluated. If a substantial subset of clear drops contains the same buffer components, the protein sample is repurified and transferred into this buffer by dialysis or spin concentration buffer exchange prior to concentration, concentrated and rescreened in this ‘optimum solubility’ buffer for crystal formation. This ‘Crystallization Optimum Solubility Screen’ is highlighted in the dark-grey boxes of Fig. 1 ▶ (middle column). We describe our use of this crystallization optimum solubility screen for the protein AF2059 (Fig. 1 ▶, dark-grey boxes in rectangular frame; right column) which, once screened for crystallization in the empirically determined optimal protein buffer, resulted in the production of crystals suitable for data collection and structure determination to 2.2 Å.
Figure 1.
Flowchart of JCSG crystal-production pipeline (light-grey boxes) and the incorporation of Crystallization Optimum Solubility Screening (dark-grey boxes) and its application to protein AF2059 (framed).
2. Experimental methods and results
2.1. Protein production and initial crystallization trials
AF2059 (TIGR AF2059; SWISS-PROT O28220) was amplified by PCR from genomic DNA from Archeaglobus fulgidus using Taq T33 polymerase (Stratagene) and primer pairs encoding the predicted 5′- and 3′-ends of AF2059. The PCR product was cloned into plasmid pMH1, which encodes an expression and purification tag consisting of MGSDKIHHHHHH at the amino-terminus of the full-length protein. The cloning junctions were confirmed by sequencing. Protein expression was performed in selenomethionine-containing medium using the Escherichia coli methionine-auxotrophic strain DL41. Bacteria were lysed by sonication in lysis buffer (50 mM K3PO4 pH 7.8, 300 mM NaCl, 10% glycerol, 5 mM imidazole, Roche EDTA-free protease-inhibitor tablets) with 0.5 mg ml−1 lysozyme. Immediately after sonication, the cell debris was pelleted by ultracentrifugation at 60 000g for 20 min (277 K). The soluble fraction was applied onto a gravity-flow metal-chelate column (Talon resin charged with cobalt; Clontech) equilibrated in lysis buffer. The column was then washed with seven column volumes (CV) of wash buffer (20 mM Tris pH 7.8, 300 mM NaCl, 10% glycerol, 10 mM imidazole) and eluted with 3 CV of elution buffer (25 mM Tris pH 7.8, 15 mM NaCl, 150 mM imidazole). Elution fractions were filtered and applied onto a Sepharose HQ POROS anion-exchange column (Boehringer Mannheim) equilibrated in anion-exchange equilibration buffer (25 mM Tris pH 7.8). The column was then washed with 3 CV of equilibration buffer and eluted with a salt gradient from 25 to 500 mM NaCl. The protein eluted in a single peak and was immediately buffer-exchanged into 10 mM Tris pH 7.8, 100 mM NaCl and concentrated to 8 mg ml−1 by centrifugal ultrafiltration (Orbital). Concentration in this buffer resulted in extensive precipitation. The protein was then centrifuged to pellet the precipitate and subjected to crystallization trials against 192 different crystallization conditions, which included the 96 conditions that constitute the ‘Core Screen’ (Page et al., 2003 ▶; Page & Stevens, 2004 ▶) and 96 PEG/Ion conditions that ranged in pH from 4.0 to 10.0 at 293 K using the nanodroplet vapor-diffusion method (Santarsiero et al., 2002 ▶).
2.2. Identification of the optimal protein buffer using initial crystallization results (Crystallization Optimum Solubility Screening)
Crystallization drops were examined manually 1, 7 and 14 d following setup. Nearly 95% of the drops (182 out of 192; most after only one day) resulted in some form of precipitation, one of which was annotated as potentially crystalline; only ten drops remained clear (Table 1 ▶). Upon examination of the conditions of the clear drops, it was observed that five of the ten clear drops contained the same buffer, 100 mM CHES pH 9.25. We concluded that this buffer was likely to improve the solubility of this protein and based on these results, AF2059 was repurified using the same protocols as those described above, with the following major difference: following elution from the anion-exchange column, the AF2059 protein was immediately buffer-exchanged into its empirically identified solubility-enhancing buffer, 100 mM CHES pH 9.25, 100 mM NaCl. In contrast to what was observed for the AF2059 protein in the original crystallization buffer (10 mM Tris pH 7.8, 100 mM NaCl), which precipitated during concentration to 8 mg ml−1, in the optimized buffer (100 mM CHES pH 9.25, 100 mM NaCl) the AF2059 protein concentrated to 16 mg ml−1 with no precipitation.
Table 1. The ten crystallization conditions that resulted in clear drops during the initial crystallization screen of AF2059.
| No. | Buffer | Precipitant |
|---|---|---|
| 1 | 100 mM CHES pH 9.25 | 5.0%(w/v) PEG 8000 |
| 2 | 100 mM CHES pH 9.25 | 5.0%(v/v) mPEG 550 |
| 3 | 100 mM CHES pH 9.25 | 10%(v/v) MPD |
| 4 | 100 mM CHES pH 9.25 | 6%(w/v) mPEG 2000 |
| 5 | 100 mM CHES pH 9.25 | 20%(v/v) MPD |
| 6 | 100 mM CHES pH 9.50 | 1.26 M (NH4)2SO4, 200 mM NaCl |
| 7 | None | 10%(w/v) PEG 1000, 10%(w/v) PEG 8000 |
| 8 | 100 mM acetate pH 4.50 | 1.0 M (NH4)2HPO4 |
| 9 | 100 mM MES pH 6.50 | 1.6 M MgSO4·7H2O |
| 10 | 100 mM Tris pH 8.50 | 50%(v/v) ethylene glycol, 200 mM MgCl2 |
2.3. Crystal formation and optimization
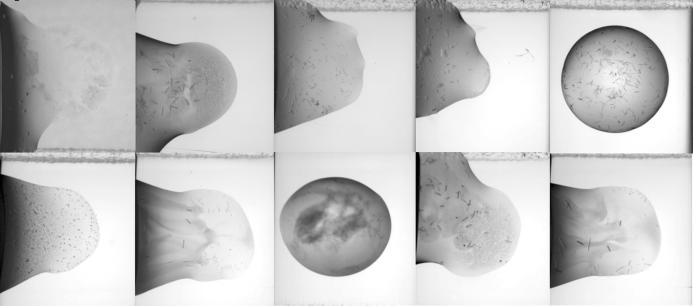
The buffer-optimized AF2059 protein was then rescreened for crystal formation against the same 192 conditions used in the initial screen at two temperatures, 277 and 293 K. In this second screen, 25% (96) of the drops were clear and 6% (24) of the drops produced crystals (Fig. 2 ▶). Several of these lead conditions were fine-screened, one of which resulted in the production of diffraction-quality crystals suitable for structure determination. The final crystallization condition that produced 2.2 Å diffraction data contained 20% MPD, 100 mM citrate pH 4.5 with 10 mM phenol, mixed with 9.5 mg ml−1 AF2059 buffered in 100 mM CHES pH 9.25, 100 mM NaCl. Data-collection and structure-determination results from this crystal will be described elsewhere.
Figure 2.
Crystallization results of protein AF2059 equilibrated in 100 mM CHES pH 9.25, 100 mM NaCl prior to crystallization trials: examples of formed crystals. Ten of the 24 drops that produced crystals when AF2059 protein equilibrated in 100 mM CHES pH 9.25, 100 mM NaCl.
3. Discussion
Inspired by the successful work of Jancarik et al. (2004 ▶), who described the development of an optimum solubility screen to identify buffer(s) for the production of monodisperse protein samples suitable for crystallization trials, we adapted this screen so that it does not require an additional step to be implemented into the structure-determination workflow. Instead of carrying out a separate optimum solubility screen prior to crystallization trials, we used the results of initial crystallization trials to guide the identification of buffers suitable for improving solubility, and in turn the crystallization potential, of a protein of interest (Fig. 1 ▶; Crystallization Optimum Solubility Screen). As a proof of principle, the A. fulgidus protein AF2059 was processed using previously published protocols for cloning, expression, purification and crystallization (Lesley et al., 2002 ▶; Levin et al., 2004 ▶). During purification, concentration to 8 mg ml−1 and initial crystallization trials, the protein precipitated heavily and only 5% of crystallization conditions screened resulted in clear drops (Table 1 ▶). Since the presence of ‘clear drops’ was the first criteria used by Jancarik et al. (2004 ▶) to identify the optimal buffer(s) for the production of monodisperse protein samples suitable for crystallization trials, we decided to buffer-exchange AF2059 into the ‘clear-drop’ buffer of 100 mM CHES pH 9.25, 100 mM NaCl. This allowed us to concentrate the protein to 16 mg ml−1 with no precipitation. This ‘buffer-optimized AF2059’ sample was then screened against the same 192 crystallization conditions and resulted in 24 crystal leads. Iterative fine-screening about several of these lead conditions resulted in the production of diffraction-quality crystals suitable for structure determination and the structure was solved to 2.2 Å (Fig. 2 ▶; data collection and structure determination will be described elsewhere).
These results show that crystallization screens can be used not only to identify conditions that lead to the formation of diffraction-quality crystals, but also to identify buffers that can improve the solubility of the protein and in turn improve the likelihood that it will crystallize. Specifically, if initial crystallization trials fail to produce crystals, the results can be examined again to identify the clear drops, i.e. those which increase the solubility of the protein. Once a buffer or set of buffers has been identified, the protein can be exchanged into these buffer solutions and crystallization trials repeated with the buffer-optimized protein. Alternatively, the buffer-optimized protein can be further analyzed using dynamic light scattering to determine its aggregation state (Jancarik et al., 2004 ▶). The latter was not implemented in this study because we wanted to develop a method that would have the smallest impact on the workflow of the JCSG structure-determination pipeline. Benefits of this Crystallization Optimum Solubility Screen are the following. Firstly, hundreds of conditions (crystallization conditions) can be screened simultaneously with very small volumes of protein (nanolitre volumes), enabling a larger set of buffers to be sampled simultaneously. Secondly, if the crystallization results are recorded electronically, the images can be analyzed using automated programs for crystal identification to identify the clear drops, the components of those crystallization conditions listed and ranked for the components present most frequently. Using this Crystallization Optimum Solubility Screen, optimum solubility buffers can be rapidly identified, enabling the protein to be exchanged into the buffer(s) identified most frequently and immediately rescreened for improved crystal formation.
Acknowledgments
The authors thank the members of the Joint Center for Structural Genomics for target selection and structure determination. This work was supported by the Protein Structure Initiative 1 NIGMS grant to the JCSG (GM62411).
References
- D’Arcy, A. (1994). Acta Cryst. D50, 469–471. [DOI] [PubMed] [Google Scholar]
- Jancarik, J., Pufan, R., Hong, C., Kim, S.-H. & Kim, R. (2004). Acta Cryst. D60, 1670–1673. [DOI] [PubMed] [Google Scholar]
- Lesley, S. A. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 11664–11669. [Google Scholar]
- Levin, I. et al. (2004). Proteins, 56, 404–408. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1994). Crystallization of Biological Macromolecules. Cold Spring Harbor, USA: Cold Spring Harbor Laboratory Press.
- Page, R., Grzechnik, S. K., Canaves, J. M., Spraggon, G., Kreusch, A., Kuhn, P., Stevens, R. C. & Lesley, S. A. (2003). Acta Cryst. D59, 1028–1037. [DOI] [PubMed] [Google Scholar]
- Page, R. & Stevens, R. C. (2004). Methods, 34, 373–389. [DOI] [PubMed] [Google Scholar]
- Santarsiero, B. D., Yegian, D. T., Lee, C. C., Spraggon, G., Gu, J., Scheibe, D., Uber, D. C., Cornell, E. W., Nordmeyer, R. A., Kolbe, W. F., Jin, J., Jones, A. L., Jaklevic, J. M., Schutlz, P. G. & Stevens, R. C. (2002). J. Appl. Cryst., 35, 278–281. [Google Scholar]
- Wilson, W. W. (2003). J. Struct. Biol.142, 56–65. [DOI] [PubMed] [Google Scholar]