Crystallization of Arabidopsis thaliana cyclophilin 38. The crystal diffracts X-rays to 2.5 Å resolution.
Keywords: cyclophilins, PPIases, AtCyp38
Abstract
AtCyp38 is one of the highly divergent multidomain cyclophilins from Arabidopsis thaliana. A recombinant form of AtCyp38 (residues 83–437) was expressed in Escherichia coli and purified to homogeneity. The protein was crystallized using the vapour-batch technique with PEG 6000 and t-butanol as precipitants. Crystals of recombinant AtCyp38 diffracted X-rays to better than 2.5 Å resolution at 95 K using a synchrotron-radiation source. The crystal belongs to the C-centred orthorhombic space group C2221, with unit-cell parameters a = 58.2, b = 95.9, c = 167.5 Å, and contains one molecule in the asymmetric unit. The selenomethionine derivative of the AtCyp38 protein was overexpressed, purified and crystallized in the same space group and data were collected to 3.5 Å at the NSLS synchrotron. The structure is being solved by the MAD method.
1. Introduction
The proteins that have been identified as the cellular receptors of immunosuppressant drugs have been aptly termed immunophilins. Immunophilins are present in organisms ranging from bacteria to animals. They are characterized by their enzymatic activity: the peptidyl-prolyl cis–trans isomerization of polypeptides. Even though peptidyl-prolyl cis–trans isomerase (PPIase) activity is an important function assigned to this group of proteins that is responsible for proper protein folding, immunophilins also possess several other functions (Romano et al., 2005 ▶). They are classified into two major families according to their binding partners, namely FK506-binding proteins (FKBPs) and cyclosporin-binding proteins (cyclophilins).
Immunophilins form a significant component of the chloroplast proteome in plants. Their abundance in the thylakoid lumen suggests that these proteins may play important roles in this relatively uncharacterized subcellular compartment. Moreover, the importance of some of the complex multidomain immunophilins in functions pertaining to development is underscored by the strong phenotypes displayed by their corresponding mutants (Romano et al., 2005 ▶). The evidence obtained so far suggests that they may also be involved in plant development and photosynthesis, mainly owing to their additional functional domains. The plant immunophilin family consists of a wide variety of isoforms varying in form, function and cellular location. The Arabidopsis genome-sequencing project has allowed the identification of 29 isoforms of cyclophilins alone (Romano et al., 2004 ▶). The multidomain cyclophilin isoforms encompass a set of proteins possessing unique domain arrangements. AtCyp38 is one of the highly divergent multidomain cyclophilins from A. thaliana. It possesses an atypical poorly conserved primary structure and shows only very little similarity to human cyclophilin A, on which several structural studies have been carried out. AtCyp38 is composed of an N-terminal leucine-zipper domain, a central acidic region and the C-terminal catalytic PPIase (cyclophilin) domain (He et al., 2004 ▶; Romano et al., 2004 ▶). All other known cyclophilin structures, except that of bovine Cyp40, have a single-domain organization. Bovine Cyp40, although a multidomain cyclophilin, has its cyclophilin domain in the N-terminus and a C-terminal TPR triplet separated by two putative nuclear targeting signals (Taylor et al., 2001 ▶).
AtCyp38, along with other lumenal cyclophilins, may have forfeited its PPIase function in order to carry out novel roles within the catalytically suboptimal cellular environment. A homologue of AtCyp38, Tlp40, has previously been identified from the spinach lumen (Fulgosi et al., 1998 ▶). This protein associates with and regulates the activity of a membrane phosphatase involved in protein dephosphorylation at the Photosystem II (PS II) reaction centre (Vener et al., 1999 ▶). Following an abrupt elevation in temperature, Tlp40 is released from the thylakoid membrane into the lumen, coinciding with phosphatase activation and dephosphorylation of PS II reaction-centre proteins (Rokka et al., 2000 ▶). The localization of AtCyp38 suggests that it might have evolved with new functions.
So far, no structure is available for the family of plant cyclophilins. The exact functions of these proteins can only be appreciated through their three-dimensional structures, followed by a thorough functional analysis based on clues from the structures. With this goal in mind, we have undertaken the crystal structure determination of the multidomain plant cyclophilin AtCyp38. Here, we report the overexpression, purification, crystallization and preliminary X-ray results of AtCyp38.
2. Materials and methods
2.1. Protein expression and purification
The gene encoding residues 83–437 of the AtCyp38 protein was cloned into the XbaI/XhoI sites of the pGEX-KG vector and the GST-fusion protein was overexpressed in Escherichia coli BL21 (DE3) cells. In brief, the cells were grown in LB medium containing 10 µg ml−1 ampicillin to an OD600 of 0.6 at 303 K and expression of the recombinant protein was induced with 0.4 mM IPTG. Post induction, cells were grown for 4 h at 298 K and then harvested by centrifugation at 4200g for 10 min. The cell pellet was suspended in ice-cold lysis buffer containing 50 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and subjected to sonication. The crude lysate was centrifuged at 42 400g for 45 min at 277 K and the cell debris was discarded. The supernatant was applied onto a GST-affinity column and the fusion protein was allowed to bind to the resin overnight at 277 K. The contaminant proteins that were loosely bound to the affinity column were removed by alternate washes with high-salt wash buffer [20 mM Tris–HCl (pH 7.5), 900 mM NaCl, 1 mM DTT, 1 mM Na EDTA] and low-salt wash buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 1 mM Na EDTA]. The GST tag was removed from the fusion protein by overnight on-column cleavage at 277 K using the thrombin protease (Amersham Biosciences) in the low-salt buffer. The cleaved protein was further purified by gel-filtration chromatography on a HiLoad 16/60 Superdex-75 column (Amersham Biosciences) which was previously equilibrated with low-salt wash buffer containing 1 mM PMSF. The cleaved protein has an additional 14 residues from the fusion partner attached to the N-terminus. The purified protein was analyzed by SDS–PAGE, native PAGE and MALDI–TOF mass spectrometry to confirm its purity and homogeneity. Dynamic light-scattering data showed that the protein exists as a monomer. The molecular weight of AtCyp38 was determined to be 40 395 ± 1.05 Da and the protein that was used for crystallization had purity of greater than 95% (Fig. 1 ▶). The protein was stored in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM Na EDTA, 1 mM PMSF at a concentration of 5 mg ml−1 at 193 K and used for crystallization when required.
Figure 1.
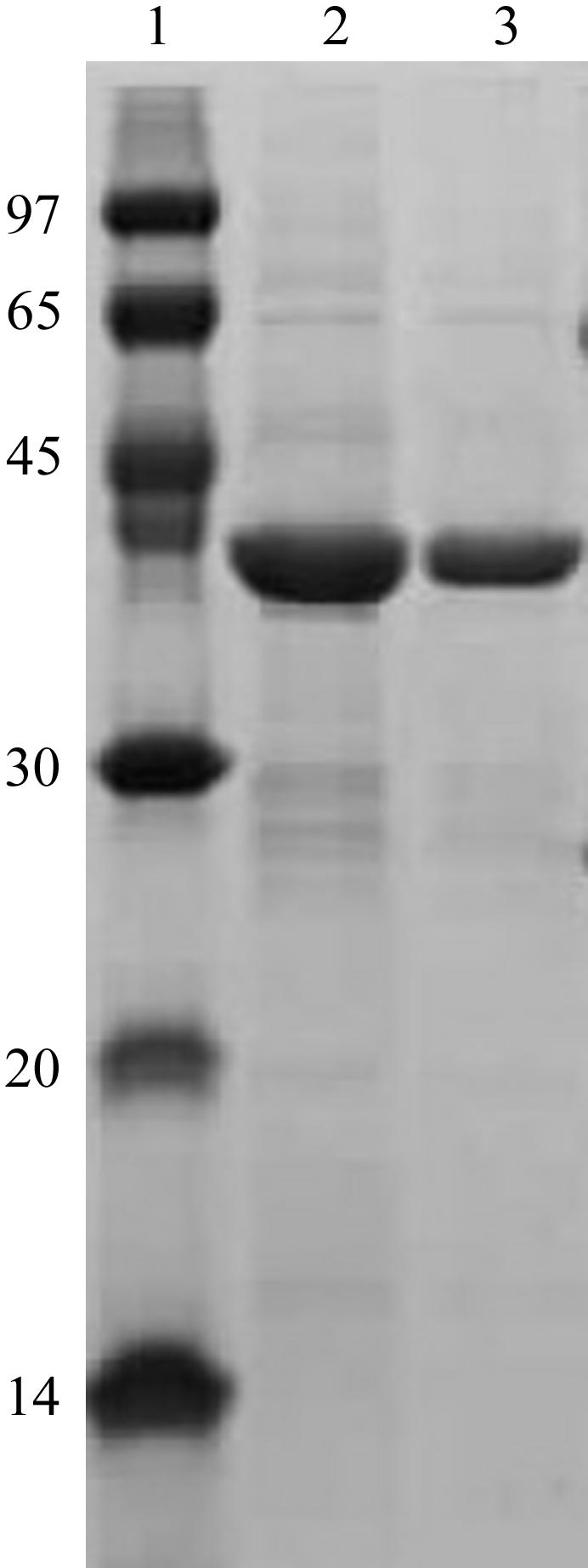
SDS–PAGE of recombinant native AtCyp38. Lane 1, molecular-weight markers (kDa); lanes 2 and 3, purified recombinant AtCyp38.
Overexpression of the selenomethionine derivative of AtCyp38 was achieved in E. coli BL21 (DE3) cells (Doublié, 1997 ▶). Cells were grown in Minimal M9 medium to an OD600 of 0.8 and the following amino acids were added to the concentrations given in parentheses: l-lysine (100 mg ml−1), l-phenylalanine (100 mg ml−1), l-threonine (100 mg ml−1), l-isoleucine (50 mg ml−1), l-leucine (50 mg ml−1), l-valine (50 mg ml−1) and l-selenomethionine (40 mg ml−1). Induction, extraction, purification, storage and analysis of the selenomethionine protein followed a similar protocol to that employed for the native protein. The mass of the selenomethionine-derivatized AtCyp38 protein from MALDI–TOF MS is 40 839 ± 1.08 Da, which suggests that approximately nine methionine residues have been replaced by selenomethionine.
2.2. Crystallization
Crystallization trials were initially performed using the vapour-diffusion method at 293 K with various commercially available crystallization kits. Crystals grew within 2 d in Hampton Crystal Screen 1 (Hampton Research) condition No. 41 [20%(w/v) PEG 4000, 10%(v/v) 2-propanol and 100 mM HEPES pH 7.5] and also in the flexible sparse-matrix screen (Zeelen, 1999 ▶) condition 2D3 consisting of 20%(w/v) PEG 6000, 2.5% t-butanol and 100 mM sodium citrate pH 5.5. Owing to the volatile nature of 2-propanol and t-butanol, the crystals disintegrated as soon as the cover slip was opened. Furthermore, these crystals did not diffract at all. The vapour-batch method (Mortuza et al., 2004 ▶) was also attempted: 2 µl AtCyp38 in 150 mM NaCl, 20 mM Tris–HCl pH 7.5, 1 mM DTT, 1 mM Na EDTA, 1 mM PMSF (protein concentration 5 mg ml−1) was mixed with an equal volume of crystallization solution containing 20%(w/v) PEG 4000, 100 mM HEPES pH 7.5 and also 20%(w/v) PEG 6000, 100 mM sodium citrate pH 5.5 in two separate vapour-batch plates. 2 µl droplets were dispensed into 96-well vapour-batch plates (Douglas Instruments) and each droplet was covered with 8 µl paraffin oil (Fluka). 5%(v/v) aqueous 2-propanol was placed in the side wells of the first tray and 1.25%(v/v) aqueous t-butanol in the side wells of the second tray. The drops were then equilibrated overnight at 293 K. Next day, the concentrations of 2-propanol and t-butanol were increased to 10 and 2.5%, respectively, and maintained at 293 K. Crystals of AtCyp38 protein and its selenomethionine derivative grew within 2 d. Further optimization of these conditions yielded crystals of approximate dimensions 0.2 × 0.1 × 0.1 mm (Fig. 2 ▶). AtCyp38 crystals from the 2-propanol condition diffracted more weakly compared with those from the t-butanol condition.
Figure 2.
A crystal of native AtCyp38 obtained by the vapour-batch method.
2.3. Data collection and analysis
Single crystals were transferred to fresh crystallization solution supplemented with 1.25% t-butanol and 20% glycerol as a cryoprotectant and then flash-cooled in liquid nitrogen at 93 K. Native and selenomethionine-derivative crystal data sets were collected at the X25 beamline, National Synchrotron Light Source, Brookhaven National Laboratory (Upton, NY, USA) with an ADSC Q315 charge-coupled device detector (Area Detector Systems Corporation). The data-collection statistics are given in Table 1 ▶.
Table 1. Crystal parameters, data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell (native, 2.59–2.50 Å; SeMet, 3.63–3.50 Å).
| SeMet | ||||
|---|---|---|---|---|
| Native | Inflection | Peak | Remote | |
| Crystal parameters | ||||
| Space group | C2221 | C2221 | ||
| Unit-cell parameters | ||||
| a (Å) | 58.2 | 58.1 | ||
| b (Å) | 95.9 | 96.0 | ||
| c (Å) | 167.5 | 167.2 | ||
| No. of molecules in ASU | 1 | 1 | ||
| Data collection | ||||
| X-ray source and detector | BNL (X25)/ADSC Q315 CCD | |||
| Resolution (Å) | 2.5 | 3.5 | ||
| Total observations | 109448 | 38377 | 38181 | 37311 |
| Unique reflections | 16710 | 6213 | 6259 | 6181 |
| Completeness (%) | 97.8 (99.8) | 98.4 (93.3) | 98.2 (91.9) | 98.3 (92.3) |
| Redundancy | 6.7 (6.7) | 6.3 (6.0) | 6.2 (5.7) | 6.1 (5.6) |
| Rsym† | 0.071 (0.29) | 0.059 (0.09) | 0.065 (0.10) | 0.050 (0.07) |
| 〈I/σ(I)〉 | 21.30 (5.4) | 30.2 (18.3) | 30.7 (17.5) | 20.5 (12.1) |
R
sym = 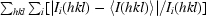 .
.
All data sets were processed and scaled using the HKL2000 program package (Otwinowski & Minor, 1997 ▶). The Matthews coefficient of the crystal (Matthews, 1968 ▶) indicated a solvent content of 57% and the asymmetric unit contains one molecule. The structure is being solved by the three-wavelength MAD method using the selenomethionine data.
References
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed] [Google Scholar]
- Fulgosi, H., Vener, A. V., Altschmied, L., Herrmann, R. G. & Andersson, B. (1998). EMBO J.17, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Li, L. & Luan, S. (2004). Plant Physiol.134, 1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mortuza, G. B., Haire, L. F., Stevens, A., Smerdon, S. J., Stoye, J. P. & Taylor, I. A. (2004). Nature (London), 431, 481–485. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rokka, A., Aro, E.-M., Herrmann, R. G., Andersson, B. & Vener, A. V. (2000). Plant Physiol.123, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, P., Gray, J., Horton, P. & Luan, S. (2005). New Phytol.166, 753–769. [DOI] [PubMed] [Google Scholar]
- Romano, P., Horton, P. & Gray, J. E. (2004). Plant Physiol.134, 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P., Dornan, J., Carrello, A., Minchin, R. F., Ratajczak, T. & Walkinshaw, M. D. (2001). Structure, 9, 431–438. [DOI] [PubMed] [Google Scholar]
- Vener, A. V., Rokka, A., Fulgosi, H., Andersson, B. & Herrmann, R. G. (1999). Biochemistry, 38, 14955–14965. [DOI] [PubMed] [Google Scholar]
- Zeelen, J. P. (1999). Protein Crystallization Techniques, Strategies and Tips. A Laboratory Manual, edited by T. Bergfors, pp. 79–90. La Jolla, CA, USA: International University Line.



