Summary
PML nuclear bodies (NBs) are nuclear structures that have been implicated in processes such as transcriptional regulation, genome stability, response to viral infection, apoptosis and tumor suppression. PML has been found to be essential for the formation of the NBs, as these structures do not form in Pml null cells, while PML add back fully rescues their formation. However, the basis for such a structural role of PML is unknown. We demonstrate that PML contains a SUMO binding motif that is independent of its SUMOylation sites and is surprisingly necessary for PML-NB formation. We demonstrate that the PML RING domain is critical for PML SUMOylation and PML-NB formation. We propose a model for PML-NB formation whereby PML SUMOylation and non-covalent binding of PML to SUMOylated PML through the SUMO binding motif constitutes the nucleation event for subsequent recruitment of SUMOylated proteins and/or proteins containing SUMO binding motifs to the PML-NBs.
Introduction
The PML tumor suppressor gene was cloned at the breakpoint of the t(15;17) chromosomal translocations of acute promyelocytic leukemia (APL) where it fuses to the retinoic acid receptor α gene (RARα) leading to the generation of an oncogenic PML-RARα chimeric protein (Pandolfi, 2001). The PML gene encodes multiple isoforms, which are derived from alternative mRNA splicing. PML nuclear isoforms are localized in distinct subnuclear structures known as PML-nuclear bodies (PML-NBs), PML oncogenic domains (POD), nuclear domain 10 (ND10), or Kremer (Kr) bodies (Hofmann and Will, 2003; Jensen et al., 2001; Zhong et al., 2000b), although cytosolic isoforms of PML have also been characterized and found to be functionally relevant (Bernardi and Pandolfi, 2003; Lin et al., 2004). The PML-NB is disrupted in APL blasts as a consequence of the dominant negative action of the PML-RARα fusion protein over PML through direct physical interaction, while PML-NB reorganization correlates with biological and clinical response upon treatment with retinoic acid (Koken et al., 1994). PML has multiple critical tumor suppressive functions, which are impaired in APL cells and in cancer cells lacking PML and PML-NBs (Gurrieri et al., 2004; Salomoni and Pandolfi, 2002).
A major advance in the current understanding of the PML-NB structure is derived from a thorough characterization of its protein, rather than nucleic acid, composition, though PML-NBs make connections with surrounding chromatins (Boisvert et al., 2000). To date more than forty proteins involved in essential cellular processes such as DNA damage response and repair, apoptosis and transcriptional regulation have been found to colocalize with PML in the PML-NBs (Hofmann and Will, 2003; Jensen et al., 2001; Zhong et al., 2000b).
A correlative microscopy and electron microscopy analysis in serial thin sections revealed the donut-shaped PML-NB structure (Boisvert et al., 2000), while the PML-NB dynamically changes its morphology during the cell cycle and in response to cellular stresses (Bernardi et al., 2004; Dellaire et al., 2006; Everett et al., 1999). We and others, using Pml knockout mice and cells, established that PML is the primary essential component of PML-NB, and that PML SUMOylation is required for PML-NB formation (Ishov et al., 1999; Wang et al., 1998; Zhong et al., 2000a; Zhong et al., 2000b). Interestingly, many of the proteins found in the PML-NBs are SUMOylated (Seeler and Dejean, 2003). Furthermore, components of SUMOylation machinery are also localized in PML-NB (Seeler and Dejean, 2003). However, to date no plausible model has been proposed to explain why PML plays such an essential structural role in the formation of the PML-NB and why PML needs to be SUMOylated to exert its function.
Since functional PML-NBs can be reconstituted in Pml−/− mouse cells by introducing wild type (wt), but not SUMOylation deficient PML (Ishov et al., 1999; Lallemand-Breitenbach et al., 2001; Zhong et al., 2000a), we have utilized this strategy to determine the requirements for PML-NB formation, and, on the basis of these findings, we now propose a new model for the PML-dependent nucleation and formation of the NB.
Results and Discussion
A SUMO binding motif mediates PML-SUMO1 interaction independently of SUMOylation
SUMO1 was originally identified as a PML interacting clone 1 (PIC1) by yeast two hybrid screening (Boddy et al., 1996), however PML SUMOylation was not discriminated from a possible non-covalent interaction between PML and SUMO1. We reasoned that if such non-covalent interaction between PML and SUMO1 does in fact take place it might be important for PML-NB formation. To resolve this issue, we generated a SUMOylation deficient version of PML IVa (Jensen et al., 2001) by mutagenizing lysines at positions 65, 160, 490 to arginines (a mutant which we termed PML3m; Figure 1B). We next transfected PML3m in Pml−/− mouse embryonic fibroblasts (MEFs) with or without GFP-SUMO1 and performed immunoprecipitation followed by Western blot analysis. Indeed, PML3m was still efficiently pulled down by GFP-SUMO1 (Figure 1C). Similar results were observed in reciprocal immunoprecipitations as well as in other cell types (Pml+/+ MEFs or 293T cells, see Supplementary Figure S1). These results suggested that PML retains a SUMO binding domain that is independent of its SUMOylation sites. During the course of this study, Song et al. reported a SUMO binding consensus sequence (VVVI) identified by NMR characterization of peptides from various proteins (Song et al., 2004). PML shares this consensus at position 508VVVI511 (Figure 1A), which is localized in exon 7 and thus is present in all PML isoforms except PML VIIb (Jensen et al., 2001). We thus mutated the putative SUMO binding motif from large non-polar (VVVI) to small non-polar amino acids (AAAS) in PMLwt or PML3m, or deleted this motif in the context of PML3m. These plasmids were once again cotransfected with GFP-SUMO1 in Pml−/− MEFs and immunoprecipitation and Western blot analysis were performed. We found that mutation or deletion of the VVVI sequence from PML3m abrogated the ability of GFP-SUMO1 to interact with PML3m (Figure 1D). Similar results were obtained in experiments performed in 293T cells and Pml−/− MEFs using different antibodies for immunoprecipitation and Western blot analysis (Supplementary Figure S2). The VVVI consensus in PML is therefore required for SUMO binding.
Figure 1. PML Has a SUMO Binding Motif Independent of Its SUMOylation Sites.
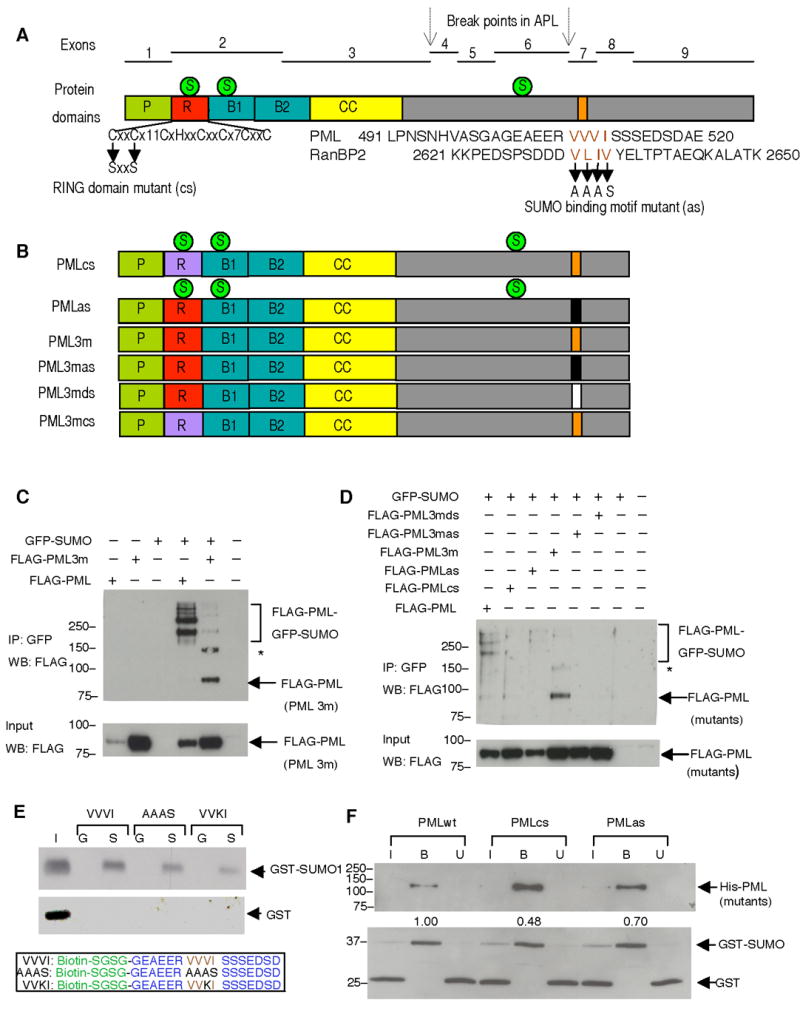
(A) Exon organization of the PML gene and the corresponding protein domains: proline rich domain (P), zinc binding boxes (B1, B2), coiled-coil domain (CC) and RING domain (R). The characteristic feature of the RING domain (Lorick et al., 1999), and two point mutations that are supposed to disrupt it, are shown below. C, cysteine, H, histidine, x, any amino acid. The numbers (11, 7) following × represent numbers of amino acids between the cysteines in PML RING domain. The putative SUMO binding motif of PML along with that of RanBP2 are aligned to reveal the consensus sequence, which is presumably composed of four bulk and hydrophobic amino acids (highlighted in brown). The VVVI in PML was mutated to AAAS in this study. The three SUMOylation sites (indicated by green circles labeled with an S), and the chromosomal break points (indicated by arrows) in acute promyelocytic leukemia (APL) are also shown. (B) Schematic presentation of PML mutants used in the study. RING domain mutant (PMLcs), SUMO binding motif mutant (PMLas), SUMOylation deficient mutant (PML3m), SUMOylation deficient and SUMO binding motif mutant (PML3mas), SUMOylation deficient and SUMO binding motif deletion mutant (PML3mds), SUMOylation deficient and RING domain mutant (PML3mcs) (C) SUMOylation deficient PML co-immunoprecipitates with GFP-SUMO1. FLAG-tagged PML or PML3m were transfected into immortalized Pml−/− MEFs with or without GFP-SUMO1. Top panel: Immunoprecipitation using anti-GFP antibodies and Western blot using anti-FLAG antibodies. Bottom panel: Western blot using anti-FLAG antibodies on 10% of protein inputs used for immunoprecipitation. (D) PML SUMO binding motif is required for SUMOylation deficient PML and SUMO co-immunoprecipitation. FLAG-tagged PML and its mutants described in (B) were analyzed for the SUMO-binding activities as in (C). Asterisk (*) indicates a cross-reacting band. Molecular weight markers (kDa) are indicated. (E) The SUMO binding motif (VVVI) in a synthetic PML-derived peptide is responsible for SUMO binding activity in vitro. Recombinant GST-SUMO1 or GST proteins were incubated with VVVI, AAAS, or VVKI peptides. The bound proteins were determined by Western blot by using antibodies against SUMO1 (upper panel), or GST (middle panel). I: input (20%), G: GST, S: GST-SUMO1. The lower panel shows peptide sequences synthesized. The biotin and linker amino acids are in green, PML sequence in blue, SUMO binding motif in brown and the mutant amino acids in the SUMO binding motif are in black. (F) PML SUMO binding motif mediates PML and SUMO interaction in vitro. Purified recombinant PMLwt, RING domain mutant (PMLcs), or SUMO binding motif mutant (PMLas) were incubated with purified GST and GST-SUMO1, the bound fractions (B) together with 10% of both input (I) and unbound (U) fractions were analyzed by SDS-PAGE and Western blot using antibodies against His (upper panel) or GST (lower panel). The numbers between panels are the ratios of GST-SUMO to PML. They are normalized to that of PMLwt, which is set to 1. Molecular weight markers (kDa) are indicated.
To determine whether the SUMO binding motif in PML directly mediated the interaction of PML with SUMO, we generated synthetic PML-derived peptides containing a wild type or mutagenized SUMO binding motif (VVVI) and tested whether they would interact with purified GST-SUMO1. Indeed, amino acid changes within the motif compromised SUMO binding activity (Figure 1E). In agreement with these findings, PMLas displayed a reduced SUMO binding activity when compared to PMLwt in in vitro binding assays utilizing purified recombinant His-tagged PMLwt or its mutants as well as GST-SUMO1 and GST (Figure 1F). Taken together, these experiments demonstrate that the SUMO binding motif in PML can directly interact with SUMO.
We also added to the analysis a PML RING domain mutant (PMLcs) in which we mutagenized the key cysteine residues to similar small non-polar amino acid serines (57C/S, 60C/S) in order to disrupt the RING structure (Figure 1A). A similar mutagenesis strategy was previously utilized to demonstrate the critical role of zinc coordinating amino acids (cysteine or histidine) in the ability of the RING finger structure to retain self-ubiquitination activity in a group of otherwise functionally unrelated proteins (Lorick et al., 1999). We observed that RING domain mutant PMLcs displayed reduced SUMO binding ability (Figure 1F). To further explore the role of RING-mediated SUMO binding, we generated a RING domain mutant in a SUMOylation deficient PML background (PML3mcs) and tested its SUMO binding activity in Pml−/− MEFs. We found that, in vivo, PML3mcs displayed a reduced interaction with SUMOylated high molecular weight proteins when compared to PML3m, but not a defect as severe as the one observed with the SUMO binding motif mutant PML3mas (Supplementary Figure S3). Collectively, our data suggest, but do not prove, that the PML RING domain can also mediate SUMO binding. We cannot rule out the possibility that the subtle nonspecific structural changes caused by RING domain mutation may result in reduced SUMO binding although we demonstrated that the RING domain mutant PML is folded properly to be able to dimerize with PMLwt in vivo (Figure 5, see below). In light of our in vivo observations (Supplementary Figure S3; Figure 5, see below), we propose that the SUMO binding motif largely contributes to interactions of PML with other SUMOylated proteins in vivo, while the SUMO binding ability of the PML RING domain would mediate a possible enzymatic activity such as PML auto-SUMOylation or the SUMOylation of other proteins, in turn, indirectly, exerting its influence in the binding of PML to other SUMOylated proteins (see below).
Figure 5. PML RING Domain Is Required for PML SUMOylation.
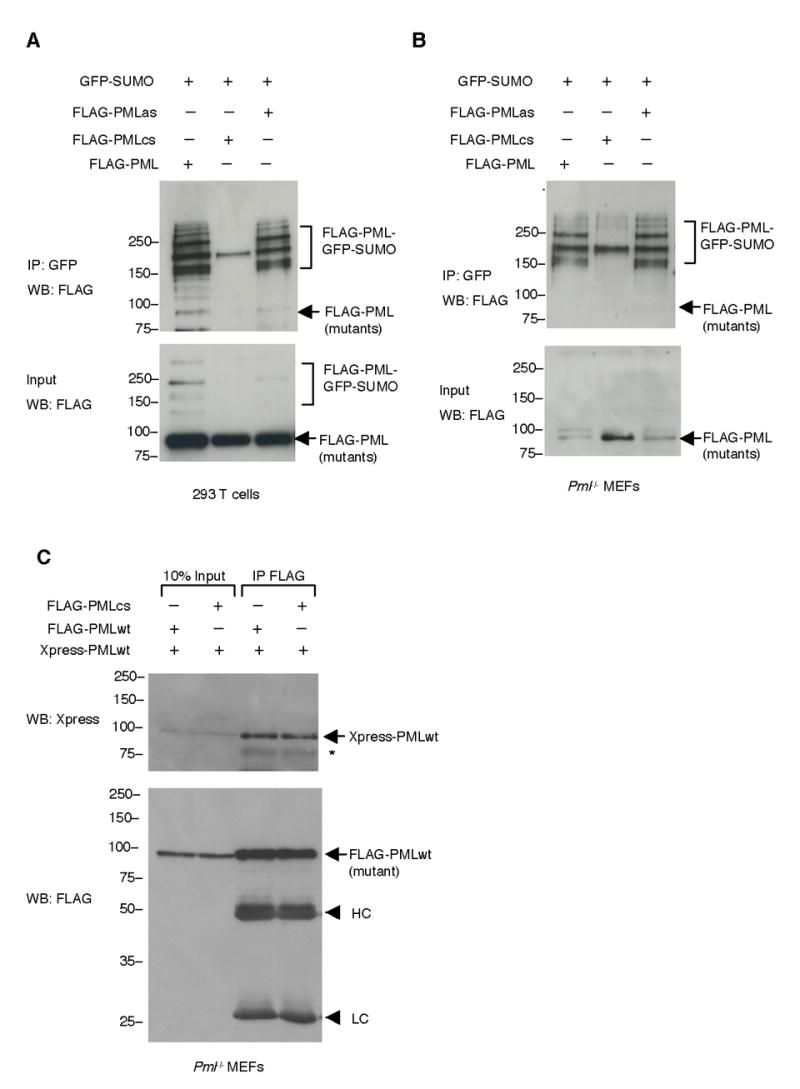
(A) Mutation of the PML RING domain drastically decreases PML SUMOylation. 293T cells were transfected with indicated plasmids. Immunoprecipitation was carried out with an anti-GFP antibody and Western blot analysis with an anti-FLAG antibody (top panel). Ten percent of the lysates used for immunoprecipitation was also analyzed by Western blot (bottom panel) (B) Pml−/− MEFs were transfected as indicated and analyzed as in (A). (C) The RING domain mutant PML maintains the ability of hetero-dimerizing with wild type PML. Pml−/− MEFs were transfected as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibodies. The immunoprecipitates and 10% of inputs used for immunoprecipitation were analyzed by Western blotting with anti-Xpress antibodies (top panel) and anti-FLAG antibodies (bottom panel). Asterisk (*) indicates an Xpress-PML degradation product. HC: Immunoglobulin heavy chains. LC: Immunoglobulin light chains. Molecular weight markers (kDa) are indicated.
PML SUMO binding motif and RING domain are required for PML-NB formation
We next tested whether the PML SUMO binding motif is required for PML-NB formation. To this end, plasmids expressing PML and the PML mutants described above were cotransfected with GFP-SUMO1 in Pml−/− MEFs and confocal immunofluorescence analysis was performed (Figure 2). As expected, cells transfected with PMLwt contained 10–20 PML-NBs per cell nucleus of heterogeneous sizes, and PMLwt was found to completely colocalize with GFP-SUMO1. PMLcs displayed predominantly a nucleoplasmic pattern, with a few dots colocalizing with GFP-SUMO1. The PML SUMO binding motif mutant (PMLas) formed fewer and aberrant structures compared to those formed by PMLwt in the nucleus, but still colocalized with GFP-SUMO1 in agreement with the notion that it can still be SUMOylated. PML3m also formed fewer aberrant compact structures compared to those formed by PMLwt in the nucleus and still colocalized with GFP-SUMO1, in agreement with the notion that it does retain the SUMO binding motif. Strikingly, however, mutagenesis or removal of the SUMO binding motif in PML3m (PML3mas, PML3mds, respectively) completely abrogated colocalization with GFP-SUMO1, although, intriguingly, PML3mas (or PML3mds) and GFP-SUMO1 were found to accumulate in adjacent subnuclear regions. By contrast, GFP-SUMO1 alone displayed a diffused nuclear staining pattern since SUMO1 mostly accumulates in PML-NBs and in Pml−/− cells PML-NBs do not form. Furthermore, we confirmed the requirement of the SUMO binding motif and RING domain in PML-NB formation by visualizing endogenous PML-NB proteins such as SUMO1 and Daxx (Supplementary Figure S4, S5). We also determined that the structures formed by PML or its mutants are distinct from nucleoli or splicing speckles (Figure 3; Figure 4). Collectively, this analysis demonstrates that the PML SUMO binding motif and an intact RING domain are required for PML-NB formation.
Figure 2. PML RING Domain and SUMO Binding Motif Are Essential for PML-NB Formation.
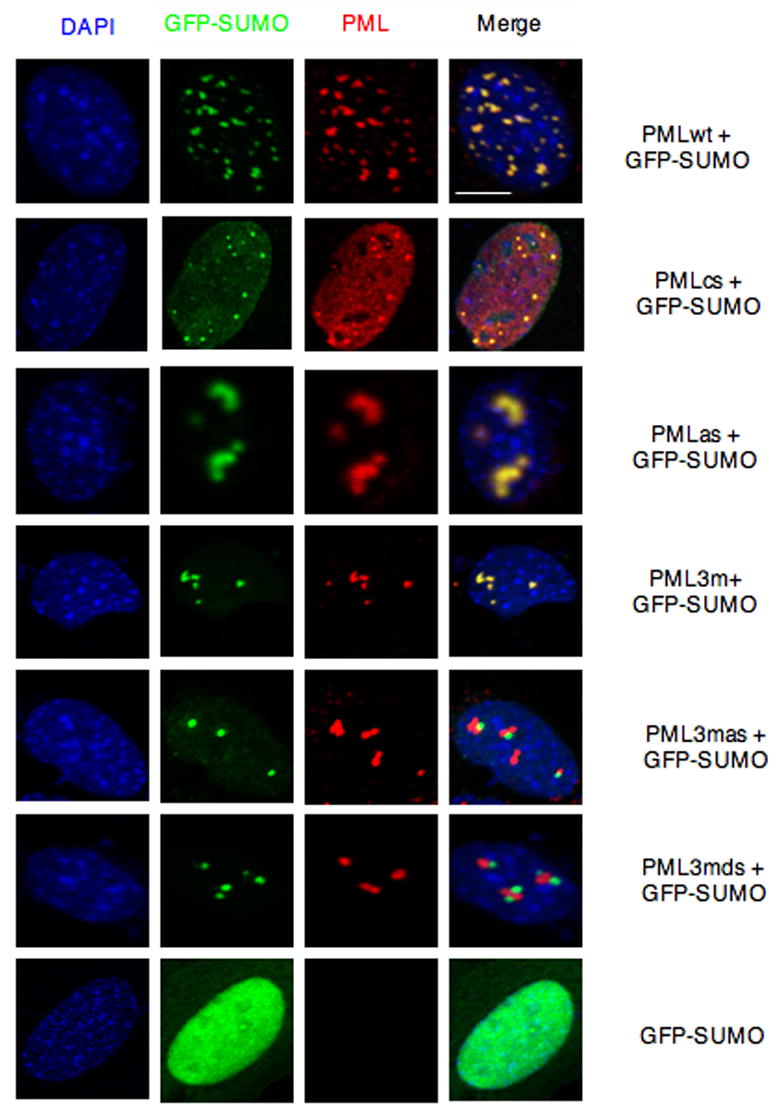
Pml−/− immortalized MEFs were transfected with the indicated plasmids and analyzed by immunofluorescence. Representative confocal microscopy images are presented. Scale bar, 10 μm.
Figure 3. The Nuclear Structures Formed by PMLwt or Its Mutants Do Not Overlap with Nucleoli.
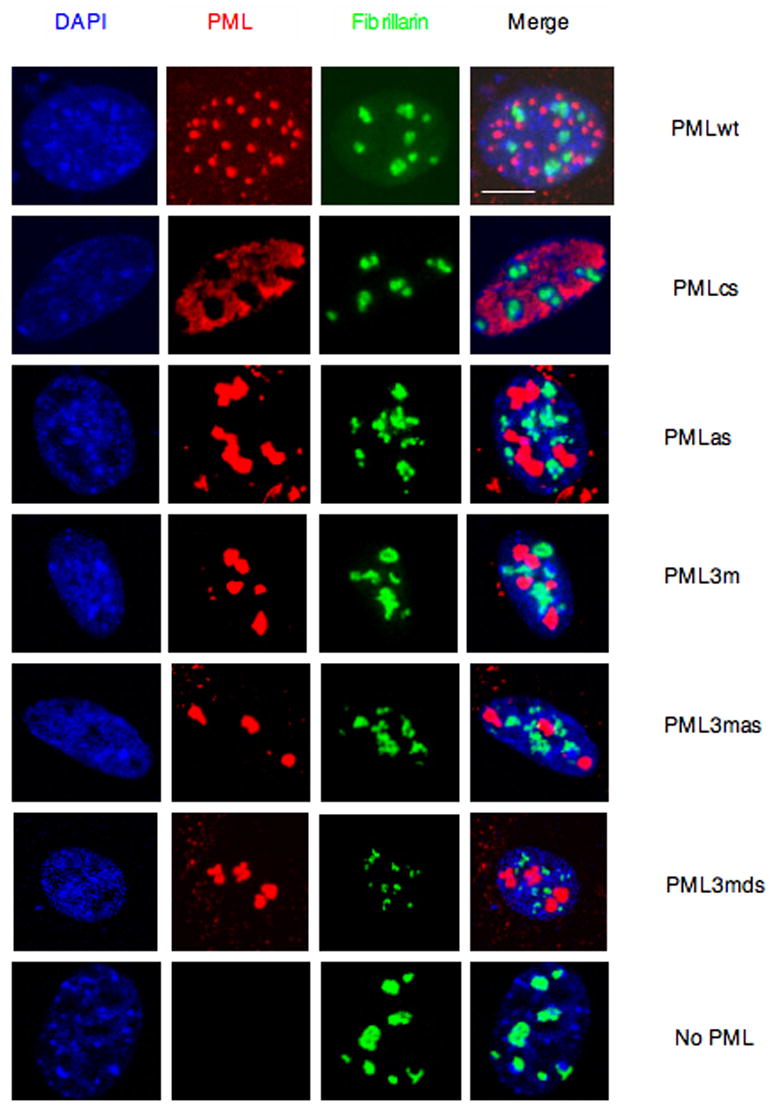
Pml−/− immortalized MEFs were transfected with the indicated plasmids and analyzed by immunofluorescence. Representative confocal microscopy images are presented. Scale bar, 10 μm.
Figure 4. The Nuclear Structures Formed by PMLwt or Its Mutants Do Not Overlap with Splicing Speckles.
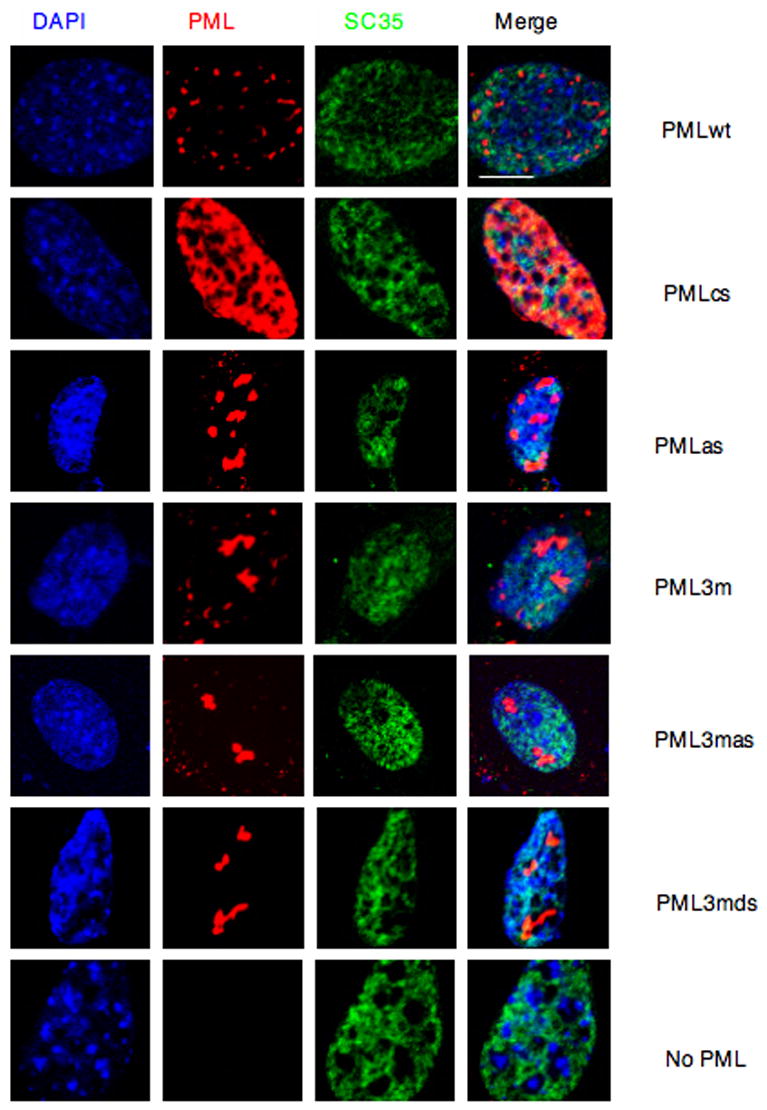
Pml−/− immortalized MEFs were transfected with the indicated plasmids and analyzed by immunofluorescence. Representative confocal microscopy images are presented. Scale bar, 10 μm.
PML RING domain is required for efficient PML SUMOylation
To explore the mechanism underlying the involvement of the PML RING domain in PML-NB formation, we first compared the levels of SUMOylation of the PML RING mutant, PMLcs, to those of PMLwt or the PML SUMO binding motif mutant, PMLas. Co-transfecting these mutants with GFP-SUMO1 and then performing immunoprecipitations to enrich the SUMOylated proteins, we found that the SUMOylation of PMLcs was drastically reduced while SUMOylation of PMLas was almost normal when compared to PMLwt (Figure 5A and 5B). To rule out the possibility that RING mutagenesis impaired PML homo-dimerization ability, PMLwt and PMLcs differently tagged (PMLwt Xpress-tagged or FLAG-tagged, and PMLcs FLAG-tagged) were cotransfected in Pml−/− MEFs and their ability to interact was assessed by immunoprecipitation and Western blot analysis. The result indicated that PMLcs displays (homo)dimerization abilities similar to the ones of PMLwt (Figure 5C). Because the RING domain is essential for the activity of a class of SUMO E3 ligases (Seeler and Dejean, 2003), and only PML (or its mutants) and GFP-SUMO1 were over-expressed in Pml−/− MEFs in these experiments, it is tempting to speculate that PML acts as a SUMO E3 ligase that catalyzes self-SUMOylation. Indeed, in agreement with this possibility, the PML RING domain was shown to be essential for PML self-SUMOylation in budding yeast, where PML or its RING domain mutant (72C/A) and SUMO1 were over-expressed (Quimby et al., 2006).
A new model for the nucleation of the PML-NB: essential role for PML SUMOylation and subsequent non-covalent binding through the SUMO binding motif
Based on our biochemical and cytomorphological analysis, we propose a new model for the formation of the PML-NB (Figure 6). In mitosis, PML is found in aggregates. This is due to its de-SUMOylation, subsequent disassembly of the PML-NBs (Everett et al., 1999), and PML dimerization/multimerization mediated by its RBCC motif (Jensen et al., 2001). In interphase, PML is SUMOylated and, importantly, once PML is SUMOylated, the PML SUMO binding motif can now mediate interactions with nearby SUMOylated PML molecules hence allowing the formation of orderly PML networks. Since a large number of proteins associated with the PML-NB are SUMOylated and/or do contain SUMO binding motifs (e.g. Daxx; see the accompanying paper by Lin et al.), it is possible that these proteins are recruited to the PML networks to eventually form higher order of protein structures such as the PML-NB through non-covalent interactions mediated by covalently bound SUMO and SUMO binding motifs present in PML and these other NB components. In agreement with this model and with the essential roles for PML SUMOylation and subsequent non-covalent binding through the SUMO binding motif, an unSUMOylatable PML is found in aggregates in the interphase nucleus, while a PML mutant that can be efficiently SUMOylated, but does not retain a functional SUMO binding motif, cannot form normal PML-NB either. This is in agreement with the notion that in both cases the formation of higher order of protein structures such as the PML-NB is prevented.
Figure 6. A Model of PML-NB Formation.
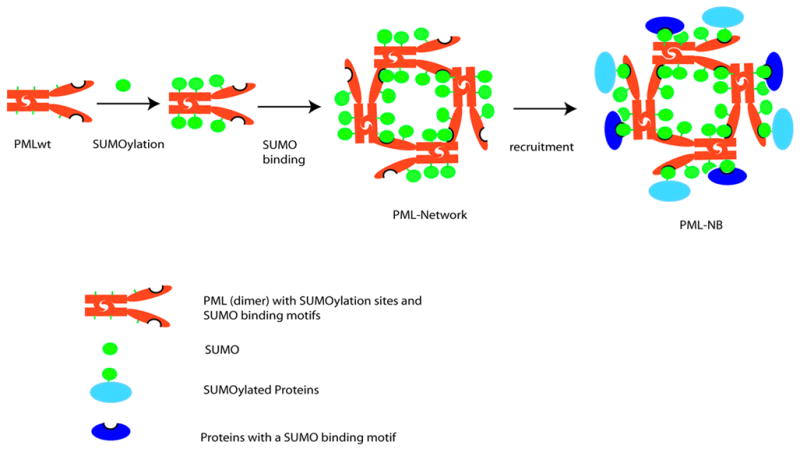
In mitosis, deSUMOylation of PML causes disassembly of PML-NBs. As a result, PML is in aggregate mediated by its RBCC motif (shown as a dimer). In interphase, PML is SUMOylated. Subsequently, the non-covalent interactions between two PML molecules mediated both by SUMO binding motif and RBCC motif promote the growth of concentered PML networks, in which nuclear body proteins that are SUMOylated and/or contain SUMO binding motifs will be recruited through SUMO moieties and SUMO binding motifs that are amply provided by PML. Although, based on this model, it is not clear how the electron dense core material in the center of the PML-NB is packaged during the NB biogenesis (not shown in the model), it is likely that PML SUMOylation is associated with the packaging process, and that the diameter of the NB is determined by the volume of the core material. Red rods: PML dimers. Green circles: SUMO. Light blue circle joined to green circle: SUMOylated proteins. Dark blue circle with indentation: proteins containing a SUMO binding motif
Biological and pathological implications of SUMOylated PML and PML SUMO binding motif-mediated non-covalent interaction
We found that SUMO binding motif or RING domain PML mutants are defective in apopotosis when expressed in Pml−/− MEFs (Supplementary Figure S6), suggesting that the tumor suppressive activity of PML may depend, at least in part, on its ability to mediate the proper formation of the PML-NBs, an idea that can now be thoroughly tested in PML mutant knock-in mice.
Our findings have implications for the pathogenesis of APL. As mentioned above, the PML SUMO binding motif is localized in PML exon 7, which is present in most PML isoforms and is invariably lost in the PML-RARα oncoprotein of APL as the chromosomal translocation breakpoints always lay in 5′ introns (3 and 6 respectively; see also Figure 1A). Loss of SUMO binding motif in the PML-RARα fusion protein may explain why the fusion protein fails to aggregate in NB-like structures (e.g. when transfected in Pml−/− MEFs, data not shown), acting instead as a dominant negative PML mutant in triggering the disruption of the PML-NB in the APL blast (Koken et al., 1994).
Similar SUMO binding motifs may be present in many proteins involved in SUMOylation related processes (Hannich et al., 2005; Minty et al., 2000; Reverter and Lima, 2005; Song et al., 2004). From a biological stand point, it is tempting to speculate that the non-covalent interaction mediated by SUMO binding motifs, by allowing protein-protein interaction to occur, may represent a general mechanism that could modulate a number of diverse functions, including the assembly of macromolecular structures such as PML-NBs, as well as the SUMOylation process itself, in analogy with ubiquitin/substrate non-covalent interactions (Gill, 2004).
Experimental Procedures
Plasmid Construction
PML used in the study was classified as PML IVa (Jensen et al., 2001). It was cloned into BamH I and EcoR I sites of pCMVtag2B vector (Stratagene) so that a FLAG tag was placed at the PML N-terminus. PML3m, PMLcs, PMLas, PML3mas, and PML3mds were constructed from the cloned FLAG-PML IVa plasmid using a Quikchange Site-Directed Mutagenesis Kit (Stratagene). Plasmids expressing His-tagged PML I mutants were generated by using a Quikchange mutagenesis based on pET33b-PML I from S. Bergmann. The plasmid expressing recombinant GST-SUMO1 was constructed by insertion of a PCR product of activated SUMO1 (C-terminal ended as two glycines) into BamH I and Xho I sites of pGEX-5X-1 (Amersham Biosciences). Retroviral vectors that express FLAG-PMLwt and FLAG-PML3m were constructed by insertion of respective PCR products into BamH I and EcoR I sites of a pBABE vector, and other mutants were subsequently made by Quikchange mutagenesis. All plasmids were verified by DNA sequencing. Primer sequences are available upon request. The pGFP-SUMO1 plasmid was a kind gift of P. Salomoni. Xpress-PML was previously described (Lin et al., 2004).
Cell lines, Transfections, Immunoprecipitations and Western Blotting
Human 293T cells, Pml+/+ immortalized mouse embryonic fibroblast (MEFs), and Pml−/− immortalized MEFs were maintained in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamate and antibiotics (1% v/v penicillin-streptomycin). Cells were split one day before transfection in 6 well plates (for 293T cells) or in 10 cm plates (for MEFs) so that cells reach to ~70% confluency upon transfection using effectene transfection reagent (Qiagen) according to supplier’s protocol. At 36–48h after transfection, cells were rinsed with cold phosphate buffered saline (PBS), and scraped in 1 ml of cold PBS with 10 mM N-ethylmaleimide (NEM, Sigma). Cell pellets were lysed at 4 °C in 200 μl of RIPA buffer (1% NP40, 0.5% DOC, 0.1% SDS in PBS) with freshly added 10 mM NEM, protease inhibitor (Roche, 50 ml/tablet) and 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma). The cell lysates were then sonicated on ice for 3 sec and centrifuged at maximum speed for 10 min at 4 °C in an Eppendorf centrifuge. The supernatants were mixed with 10 μl of washed and packed protein G sepharose beads (Amersham Biosciences) at 4 °C for 30 min. The cleared supernatants were then mixed with 10 μl of washed and packed protein G sepharose beads and 1 μl of anti-GFP (rabbit polyclonal antibody, Clontech), or anti-FLAG antibodies (Sigma) at 4 °C overnight. The beads were washed 4 times with RIPA buffer and the immunoprecipitates together with inputs were analyzed by 8% SDS-PAGE. The stacking gels were always saved for the detection of high molecular weight SUMOylated proteins. Western blotting was performed using anti-GFP (mouse monoclonal antibody, 1:1000, Clontech), or anti-FLAG antibodies (1:1000), visualized by enhanced chemiluminescence (Amersham Biosciences).
Recombinant Protein Purification, Peptide Synthesis and In Vitro Binding Assay
His-tagged Recombinant PML and mutants were produced in Rossetta 2(DE3)pLysS cells (Novagen) upon induction with 0.5 mM IPTG (Sigma) at 37 °C for 4 h. The proteins were purified using Talon magnetic beads (Dynal Biotech) according to the supplier’s procedure. Recombinant GST-SUMO1, and GST were produced in BL21 cells (Novagen) upon induction with 0.5 mM IPTG (Sigma) at 37 °C for 4 h. The proteinswere purified using glutathione-sepharose beads (Amersham Biosciences) according to the supplier’s protocol. The eluted proteins were dialyzed, concentrated by Centriprep 10 (Millipore), aliquotted and stored at −80 °C until use. The purity and concentration of purified proteins were analyzed by SDS-PAGE and Coomassie blue staining with known amount of bovine serum albumin (BSA) as a control. PML peptides were designed to contain an N-terminal biotin and tetrapeptide linker SGSG followed by 17 mers of PML centered at the SUMO binding motif VVVI, or its mutants in which VVVI was replaced with AAAS or VVKI. The peptides were synthesized and HPLC purified by the Protein Core facility at MSKCC.
For the in vitro binding assay using synthetic peptides, 3 μg of recombinant GST-SUMO1 or GST were incubated with 15 μg of biotinylated PML peptide or its mutant peptides at 4 °C overnight in 300 μl of interaction buffer containing 20 mM Hepes, pH 7.9, 150 mM KCl, 5 mM EDTA, 0.5 mM DTT, 0.1% (V/V) Nonidet P-40, 0.1% (W/V) BSA, 1 mM PMSF, 10% glycerol. Then 20 μl of streptavidin beads (Pierce) was added and incubated for 1 h. After three washes with PBS, the bound proteins were determined by Western blot analysis using antibodies against GST (1: 1000, Sigma) or SUMO1 (1: 500, Santa Cruz Biotechnology).
For the in vitro binding assay using full length recombinant proteins, 1 μg of purified His-tagged PML or its mutants was incubated with equivalent amounts of purified GST-SUMO1 and GST in 20 mM Hepes, pH7.5, 150 mM NaCl, 0.1% Tween-20 at 4 °C for 4 h. Then the mixtures were mixed with 10 μl of washed Talon magnetic beads with rotation at 4 °C for 1 h. The input and unbound fractions, together with the bound fractions after three washes, were analyzed by Western blot using antibodies against His (1:1000, Sigma) or GST (1:1000, Sigma). The intensities of His-tagged PML and GST-SUMO1 bands in the bound fractions were quantified using MacBAS V2.5-1 software and the ratios of GST-SUMO1 to PML calculated.
Immunofluorescence Microscopy
Pml−/− MEFs were grown in slide chambers and transfected with PML or its mutants and GFP-SUMO1 using effectene transfection reagent (Qiagen) according to the supplier’s protocol. After 36–48h, the cells were washed with PBS and fixed in 4% paraformaldehyde at RT for 20 min. The cells were then permeabilized in PBS containing 10% FCS and 0.1% Triton X-100. Rabbit anti-PML antibodies (PGM3, Santa Cruz Biotechnolgy) were added (1:500) and incubated in the same buffer at RT for 1 h, followed by incubation with goat anti-rabbit IgG conjugated with Alexa fluor 568 (Molecular Probes, 1:500), together with DAPI (0.1 μg/ml) at RT for 30 min. After washes, the slides were mounted. Images were captured by laser-scanning microscope (Leica) and processed in Adobe Photoshop 7.0. For the detection of endogenous protein markers, GFP-SUMO1 was omitted in transfections. These proteins include: SUMO1 (1:50, Santa Cruz Biotechnolgy), Daxx (1:25, Santa Cruz Biotechnolgy), Fibrillarin (1:100, Abcam), and SC35 (1:100, Abcam). They are all polyclonal antibodies and were used with anti-rabbit antibodies conjugated with Alexa fluor 488 (Molecular Probes, 1:500).
Retroviral production, infection and apoptosis analysis
Phoenix cells grown on 10 cm lysine coated plates (Biocoat Inc.) at 70% confluency were transfected with retroviral vectors expressing FLAG-PMLwt or its mutants using Lipofectamine 2000 (Invitrogen) according to the supplier’s protocol. After 48 h, the viruses containing media were collected, passed through a 0.45 μM filter, and directly used in the infection of Pml−/− MEFs for 48 h in the presence of 4 μg/ml of polybrene (Sigma). Infected cells were then selected for one week in the presence of 2.5 μg/ml of puromycin with 2–3 medium changes. The resulting cells were seeded on 6 well plates at 70% confluency. After 48 h, the cells were collected in PBS by trypsinization, together with dead cells in the medium. The cells were washed with PBS, fixed in 70% ethanol at 4 °C overnight. After washing with PBS, the cells were stained in PBS containing 2 μg/ml of propidium iodide (PI, BD Biosciences) and 0.5 μg/ml of DNase-free RNase (Roche) at RT for 1 h. The PI stained cells were subjected to Fluorescence Activated Cell Sorting (FACS) analysis. The results are presented in log scale to magnify the sub-G1 population of cells.
Supplementary Material
Acknowledgments
We thank R. Bernardi, L. Trotman, S. Bergmann, Z. Chen and other members of the Pandolfi laboratory for helpful discussion, advice and reagents. We greatly appreciate S. S. Yi and P. Tempst for help with peptide synthesis and J. Clohessy for assistance with the FACS analysis. We also thank I. Guernah for providing MEFs, T. Tong, R. Huq and other members in Molecular Cytology Core Facility for their assistance with imaging. We are grateful to L. DiSantis for editing and critical reading of the manuscript. This work was supported by the NIH/NCI grant (R01-CA74031) to P.P.P. The authors declare no conflicts of interest associated with the study. Correspondence and requests for materials should be addressed to P.P.P. (email: p-pandolfi@ski.mskcc.org)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernardi R, Pandolfi PP. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene. 2003;22:9048–9057. doi: 10.1038/sj.onc.1207106. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004 doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci. 2006;119:1034–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. J Cell Sci. 1999;112(Pt 24):4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. Embo J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- Lin DY, Chang CC, Huang YS, Chen YC, Lin TP, et al. Essential role of SUMO-interacting motif in mediating Daxx sumoylation, subnuclear localization and transcription of sumoylated transcription factors. Mol Cell. doi: 10.1016/j.molcel.2006.10.019. submitted. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP. In vivo analysis of the molecular genetics of acute promyelocytic leukemia. Oncogene. 2001;20:5726–5735. doi: 10.1038/sj.onc.1204600. [DOI] [PubMed] [Google Scholar]
- Quimby BB, Yong-Gonzalez V, Anan T, Strunnikov AV, Dasso M. The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene. 2006 doi: 10.1038/sj.onc.1209335. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000a;95:2748–2752. [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000b;2:E85–90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


