The yeast three-hybrid assay is an important tool for the detection of protein-ligand interactions in vivo and has recently been used successfully for the discovery of novel drug targets and the directed evolution of enzymes[1–6]. Schreiber and co-workers developed the first chemical inducer of dimerization (CID)3, a dimer of the immunosuppressant FK506[7]. Building from this work, a number of yeast three-hybrid systems based on different CIDs have been reported[2, 8]. We previously developed and optimized a three-hybrid system built around the small molecule CID dexamethasone-methotrexate (Dex-Mtx)[9–11]. The Dex and Mtx ligands were chosen because of their high affinities for their respective protein receptors, the rat glucocorticoid receptor and Escherichia coli dihydrofolate reductase, respectively[12, 13]. However, we hypothesized that the cross-reactivity of Mtx with endogenous DHFR in the yeast cells could impair transcription activation by Dex-Mtx in the yeast three-hybrid assay. In order to overcome this partial limitation we set out to design a CID that would selectively bind to E. coli DHFR and not to endogenous yeast DHFR. As an alternative to Mtx, we chose the DHFR inhibitor trimethoprim (TMP), which is known for its selectivity for bacterial forms of DHFR[14]. Studies have confirmed that while Mtx inhibits growth of wild type Saccrharomyces cerevisiae, TMP does not[15], suggesting that TMP could be a superior CID in yeast. Here we report the design, synthesis, and in vivo activity of Dex-TMP in the yeast three-hybrid assay.
By analogy to our dexamethasone-methotrexate system, we chose to build a heterodimeric CID using the ligand-receptor pairs dexamethasone (Dex)-rat glucocorticoid receptor (GR) and trimethoprim (TMP)-E. coli dihydrofolate reductase (DHFR). Both Dex and TMP can be modified without disrupting receptor binding, making them suitable CIDs[16–18]. Both ligands are cell-permeable and commercially available. The ligand-receptor pair Dex-GR has a KD of 5 nM and has been used successfully in yeast three-hybrid systems[9]. E. coli dihydrofolate reductase (DHFR) has a KI of 1.3 nM for inhibition by TMP[14]. We anticipated that the two interactions would be sufficiently strong to induce protein dimerization and transcription activation in the yeast three-hybrid assay.
Design & Synthesis of Dex-TMP
The design and synthesis of the Dex-TMP heterodimer is based on previous syntheses of Dex and TMP derivatives[16, 18, 19], with a linker analogous to that for the Dex-Mtx heterodimers most active in the yeast three-hybrid assay (Fig. 2C). Both ligands were coupled as their thiol derivatives to a di-iodo linker. First, Dex was converted to the carboxylic acid by oxidative cleavage using periodate and then derivatized with cystamine using standard peptide coupling reagents. The 4′-methoxy group of TMP was selectively cleaved in 48% hydrobromic acid. The resulting phenol was then derivatized with ethyl 5-bromovalerate using potassium tertbutoxide and converted to the corresponding carboxylic acid. Following saponification 2 equivalents of the TMP acid were reacted with cystamine dihydrochloride under standard peptide coupling conditions to generate the TMP disulfide. Finally, the Dex and TMP disulfides were reduced to their corresponding thiols using tri-n-butylphosphine. The thiol derivatives of the two ligands were coupled to the di-iodo linker sequentially, as for the previous Dex-Mtx CIDs. Thus, the Dex-TMP heterodimer was prepared from two components in a 10-step synthesis in 0.8% overall yield.
Figure 2.
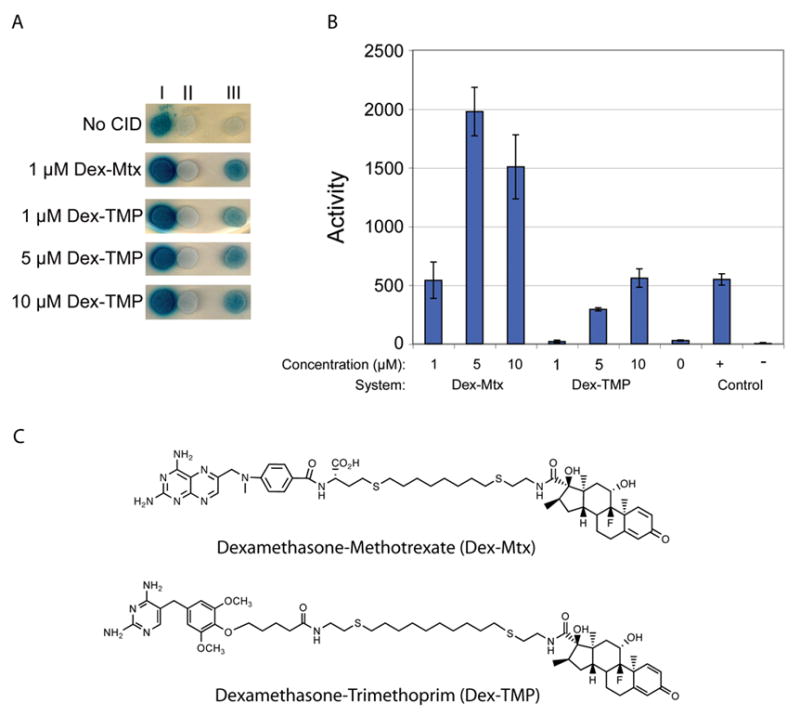
LacZ transcription assays comparing the abilities of dexamethasone-trimethoprim (Dex-TMP) and dexamethasone-methotrexate (Dex-Mtx) to activate transcription in the yeast three-hybrid assay. (A) Columns I–III on each plate correspond to yeast strains containing different DNA binding domain (DBD) and/or activation domain (AD) chimeras, and a lacZ reporter gene: I, plasmid DBD-BAIT, plasmid AD-TARGET; plasmid lacZ reporter. I is a direct protein-protein interaction used as a positive control; II, integrated DBD-DHFR, plasmid AD, plasmid lacZ reporter. II has only the AD half of the AD-GR protein chimera and is used as a negative control; III, plasmid DBD-DHFR, plasmid AD-GR, plasmid lacZ reporter. III is the plasmid based yeast three-hybrid system. Plates were grown for three days with a concentration of small molecule ranging form 1–10 μM and compared to a background of no small molecule. (B) ONPG liquid assays of CID-induced lacZ transcription. The first three bars represent the data for the all plasmid system with varying concentrations of Dex-Mtx, followed by varying concentrations of Dex-TMP and no CID. The last two rows are the positive and negative controls. (C) The structures of the two small molecules used in the study Dex-Mtx and Dex-TMP.
In Vivo Activtiy in the Yeast Three-Hybrid Assay
Dex-TMP was evaluated for its ability to activate transcription in the yeast three-hybrid assay, using a LexA DNA-binding domain-DHFR protein chimera (LexA-DHFR) and a B42 transcription activation domain-GR protein chimera (B42-GR) and a lacZ reporter gene under control of four tandem LexA operators (Fig. 1). Using standard lacZ transcription assays in both solid and liquid cultures[20], we showed that Dex-TMP can activate lacZ transcription in the yeast three-hybrid assay (Figs. 2A&B). X-gal plate assays were carried out as previously reported[10]. Yeast strains were grown on synthetic complete media lacking histidine, uracil, and tryptophan and containing no glucose, 2% galactose, 2% raffinose, and were grown with or without small molecule. In order to determine how effectively Dex-TMP activates transcription in comparison to our previous Dex-Mtx system, we tested the two CIDs side-by-side at concentrations ranging from 1–10 μM in the external growth media. Control experiments established that transcription activation was small molecule dependent, and only background levels of lacZ transcription were detected when both Dex-Mtx and Dex-TMP were omitted. Activation over background levels was observed for both small molecules and at all concentrations, except for 1 μM Dex-TMP. The maximal levels of transcription were observed with 5 μM Dex-Mtx and 10 μM Dex-TMP, respectively. In addition, all three yeast strains were grown in liquid culture and assayed for ß-galactosidase activity with ONPG as the substrate. In the liquid culture assays, growth media containing 1, 5, or 10 μM Dex-Mtx resulted in a 22, 79, and 61-fold activation over background levels, respectively. The addition of 5 and 10 μM Dex-TMP resulted in a 12 and 23-fold increase in activation, respectively.
Figure 1.
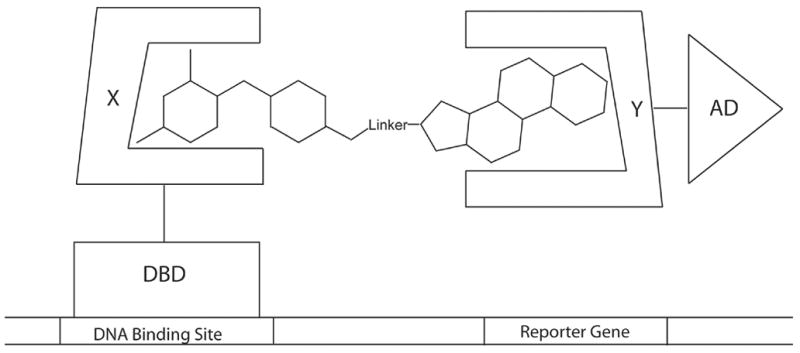
Dexamethasone-trimethoprim (Dex-TMP) yeast three-hybrid system. A heterodimeric ligand (Dex-TMP) bridges a DNA-binding domain-dihydrofolate reductase fusion protein (DBD-DHFR) and a transcriptional activator domain-glucocorticoid receptor fusion protein (AD-GR), effectively reconstituting a transcriptional activator (DBD-AD) and activating transcription of a downstream reporter gene.
Discussion
The new CID Dex-TMP can successfully dimerize the two halves of the transcriptional activator in vivo in the yeast three-hybrid assay, activating transcription of a lacZ reporter gene as shown using ß-galactosidase activity assays. Thus, the ligand receptor pair TMP-E. coli DHFR provides a new CID for use in the three-hybrid assay as well as in other in vivo applications of CIDs. This pair may prove particularly useful for applications in mammalian cell lines or even animal studies, when the toxicity of Mtx may prove problematic. However, somewhat surprisingly, the new CID does not induce transcription activation as efficiently as Dex-Mtx. There is evidence, however, that TMP-SLF activates transcription in a yeast three-hybrid assay slightly better than Mtx-SLF (unpublished data). These results point to the complexities of manipulating molecules at the cellular level. There are several plausible explanations for the difference in activity between Dex-TMP and Dex-Mtx. One possible reason for the disparity in activity is the large difference in affinities of the two for DHFR. Mtx binds E. coli DHFR with picomolar affinity (KD = ca. 10 pM)[21], whereas TMP’s affinity is much lower (KI = 1.3 nM)[14]. Also, studies have found that although yeast DHFR is not a target of TMP, the small molecule may bind to another yeast protein of unknown function[22]. Dex-Mtx may have more favorable cell permeability properties than Dex-TMP. In summary, this study provides a new CID pair, TMP-DHFR, which may be particularly advantageous for applications in mammalian cell lines and animal studies and illustrates the complexities of “engineering” at the cellular level.
Supplementary Material
- CID
chemical inducer of dimerization
- Dex-Mtx
dexamethasone-methotrexate
- Dex
dexamethasone
- Mtx
methotrexate
- GR
glucocorticoid receptor
- DHFR
dihydrofolate reductase
- TMP
trimethoprim
- Dex-TMP
dexamthasone-trimethoprim
- DBD
DNA-binding domain
- AD
transcriptional activator domain
Footnotes
We are grateful for financial support for this work from the National Institutes of Health, GM071754-01.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- 1.Lin HN, Tao HY, Cornish VW. Directed evolution of a glycosynthase via chemical complementation. Journal of the American Chemical Society. 2004;126(46):15051–15059. doi: 10.1021/ja046238v. [DOI] [PubMed] [Google Scholar]
- 2.Lin HN, V, Cornish W. In vivo protein-protein interaction assays: Beyond proteins. Angewandte Chemie-International Edition. 2001;40(5):871–875. doi: 10.1002/1521-3773(20010302)40:5<871::AID-ANIE871>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 3.Johnsson N, Johnsson K. A fusion of disciplines: Chemical approaches to exploit fusion proteins for functional genomics. Chembiochem. 2003;4(9):803–810. doi: 10.1002/cbic.200200603. [DOI] [PubMed] [Google Scholar]
- 4.Becker F, et al. A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chemistry & Biology. 2004;11(2):211–223. doi: 10.1016/j.chembiol.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Mapp AK. Regulating transcription: a chemical perspective. Organic & Biomolecular Chemistry. 2003;1(13):2217–2220. doi: 10.1039/b302656f. [DOI] [PubMed] [Google Scholar]
- 6.Hu JC. Model systems: Studying molecular recognition using bacterial n-hybrid systems. Trends in Microbiology. 2001;9(5):219–222. doi: 10.1016/s0966-842x(01)02019-4. [DOI] [PubMed] [Google Scholar]
- 7.Spencer DM, et al. Controlling Signal-Transduction with Synthetic Ligands. Science. 1993;262(5136):1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 8.Hussey SL, Muddana SS, Peterson BR. Synthesis of a beta-estradiol-biotin chimera that potently heterodimerizes estrogen receptor and streptavidin proteins in a yeast three-hybrid system. Journal of the American Chemical Society. 2003;125(13):3692–3693. doi: 10.1021/ja0293305. [DOI] [PubMed] [Google Scholar]
- 9.Lin HN, et al. Dexamethasone-methotrexate: An efficient chemical inducer of protein dimerization in vivo. Journal of the American Chemical Society. 2000;122(17):4247–4248. [Google Scholar]
- 10.Abida WM, et al. Receptor-dependence of the transcription read-out in a small-molecule three-hybrid system. Chembiochem. 2002;3(9):887–895. doi: 10.1002/1439-7633(20020902)3:9<887::AID-CBIC887>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Baker K, et al. An optimized dexamethasone-methotrexate yeast 3-hybrid system for high-throughput screening of small molecule-protein interactions. Analytical Biochemistry. 2003;315(1):134–137. doi: 10.1016/s0003-2697(02)00698-x. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborti PK, et al. Creation of Super Glucocorticoid Receptors by Point Mutations in the Steroid Binding Domain. Journal of Biological Chemistry. 1991;266(33):22075–22078. [PubMed] [Google Scholar]
- 13.Sasso SP, et al. Thermodynamic Study of Dihydrofolate-Reductase Inhibitor Selectivity. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology. 1994;1207(1):74–79. doi: 10.1016/0167-4838(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 14.Baccanari DP, Daluge S, King RW. Inhibition of Dihydrofolate-Reductase - Effect of Reduced Nicotinamide Adenine-Dinucleotide Phosphate on the Selectivity and Affinity of Diaminobenzylpyrimidines. Biochemistry. 1982;21(20):5068–5075. doi: 10.1021/bi00263a034. [DOI] [PubMed] [Google Scholar]
- 15.Game JC, Little JG, Haynes RH. Yeast Mutants Sensitive to Trimethoprim. Mutation Research. 1975;28(2):175–182. doi: 10.1016/0027-5107(75)90094-9. [DOI] [PubMed] [Google Scholar]
- 16.Roth B, et al. 2,4-Diamino-5-Benzylpyrimidines and Analogs as Anti-Bacterial Agents .5. 3',5'-Dimethoxy-4'-Substituted-Benzyl Analogs of Trimethoprim. Journal of Medicinal Chemistry. 1981;24(8):933–941. doi: 10.1021/jm00140a005. [DOI] [PubMed] [Google Scholar]
- 17.Manz B, et al. Synthesis of Biotin-Labeled Dexamethasone Derivatives - Novel Hormone-Affinity Probes. European Journal of Biochemistry. 1983;131(2):333–338. doi: 10.1111/j.1432-1033.1983.tb07266.x. [DOI] [PubMed] [Google Scholar]
- 18.Govindan MV, Manz B. 3-Step Purification of Glucocorticoid Receptors from Rat-Liver. European Journal of Biochemistry. 1980;108(1):47–53. doi: 10.1111/j.1432-1033.1980.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 19.Brossi A, et al. Synthesis and Chemotherapeutic Activity of 2 Metabolites of Trimethoprim. Journal of Medicinal Chemistry. 1971;14(1):58. doi: 10.1021/jm00283a017. [DOI] [PubMed] [Google Scholar]
- 20.Adams A, et al., editors. Methods in Yeast Genetics. Cold Spring Laboratory Press; Plainview: 1998. [Google Scholar]
- 21.Appleman JR, et al. Role of Aspartate-27 in the Binding of Methotrexate to Dihydrofolate-Reductase from Escherichia-Coli. Journal of Biological Chemistry. 1988;263(19):9187–9198. [PubMed] [Google Scholar]
- 22.Barclay BJ, Nagel MG, Huang T. Dihydrofolate reductase is not the target of trimethoprim in Saccharomyces cerevisiae. Adv Exp Med Biol. 1993;338:551–4. doi: 10.1007/978-1-4615-2960-6_112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


