Abstract
The program RIVEM (Radial Interpretation of Viral Electron density Maps) was developed to project density radially onto a sphere that is then presented as a stereographic diagram. This permits features resulting from an asymmetric reconstruction to be projected and positioned onto an icosahedral virus surface. The features that constitute the viral surface can also be simultaneously represented in terms of atoms, amino acid residues, potential charge distribution, and surface topology. The procedure can also be adapted for the investigation of various molecular interactions.
Keywords: Cryo-EM, density interpretation, asymmetry, stereographic projection
Introduction
The combination of X-ray crystallography and cryo-electron microscopy (cryo-EM) has proven to be an effective technique to analyze macromolecular assemblies (Baker et al., 1999; Chiu et al., 1999; Grimes et al., 1999; Rossmann et al., 2005). Examples include studies of various viruses (Baker et al., 1999; Böttcher et al., 1997b; Conway et al., 1997; Leiman et al., 2004; Morais et al., 2005; Zhou et al., 2000), virus/receptor complexes (Belnap et al., 2000; Bubeck et al., 2005; Hewat et al., 2000; Rossmann et al., 2002; Xiao et al., 2005a), many important cellular complexes such as ribosomes (Allen et al., 2005; Matadeen et al., 1999), nuclear pores (Beck et al., 2004), bacterial flagella (Yonekura et al., 2005), GroEL (Ludtke et al., 2004), membrane Ca2+ channels (Serysheva et al., 2005), ATPases (Bernal and Stock, 2004; Chen et al., 2004), and many large protein complexes (Acehan et al., 2002; Cheng et al., 2004; Ishikawa et al., 2004; Zhou et al., 2001). Cryo-EM has significantly improved in the last ten years to achieve sub-nanometer resolution, where secondary structural features become visible (Böttcher et al., 1997b; Conway et al., 1997; van Heel et al., 2000; Zhou et al., 2000). Better cryo-EM images can now be recorded using high voltage electron microscopes equipped with field emission guns, which provide brighter and more coherent electron beams than were previously possible. Furthermore, taking advantage of modern parallelized computer clusters, more than 103 to 105 individual particle images can be included in cryo-EM reconstructions, making it feasible to reach greatly improved resolution limits. Over the last decade, many computer programs (Baker and Cheng, 1996; Grigorieff, 1998) and software packages (Frank et al., 1996; Ludtke et al., 1999; van Heel et al., 2000) have been developed or improved with better algorithms and better user interfaces. These have facilitated the image reconstruction process to be routine and efficient. On the other hand, interpretation and visualization of the cryo-EM maps has become more difficult as the detail within higher resolution maps increases. Various programs have been developed to help analyze higher resolution cryo-EM results. These include programs for the fitting of X-ray crystallographically determined structures into cryo-EM densities (Roseman, 2000; Rossmann et al., 2001; Volkmann and Hanein, 2003; Wriggers et al., 1999) and programs for the visualization of macromolecules in cryo-EM density maps (Gillet et al., 2005; Pettersen et al., 2004). Nevertheless, interpretation of the final results of cryo-EM reconstructions can sometimes be helped by alternate methods of presentation.
Because of their exceptionally high symmetry, icosahedral viruses (Rossmann and Johnson, 1989), virus/receptor complexes (Rossmann et al., 2002), and icosahedral protein complexes (Fotin et al., 2004; Liu et al., 2004; Walz et al., 1999) have been successfully analyzed by X-ray crystallography and cryo-EM. However, recent studies have shown that some macromolecular assemblies that had been assumed to be icosahedral do not have perfect symmetry or have their symmetry broken during certain stages of their life cycle (Rossmann et al., 2006). Some examples are tailed bacterial phages, which have incomplete icosahedral symmetry due to the attachment of a tail (Cerritelli et al., 2003; Jiang et al., 2006; Lander et al., 2006; Morais et al., 2005; Orlova et al., 2003); nucleocytoplasmic large DNA viruses, which also can have a unique vertex (Van Etten et al., 1991; Xiao et al., 2005b); and some parvoviruses, which can attach their receptors in an asymmetric manner (Hafenstein et al., 2006). In order to locate the position of asymmetrically distributed densities in an icosahedral virus capsid, a program (Radial Interpretation of Viral Electron density Maps or RIVEM) was developed for projecting the asymmetric density in the context of the assumed symmetry axes onto a spherical surface.
The surfaces of icosahedral viruses (Kolatkar et al., 1999; Rossmann et al., 2002) have been conveniently displayed as “roadmaps” (Chapman, 1993; Rossmann and Palmenberg, 1988). However, the earlier roadmap programs, which projected the viral surface onto a plane, had limitations that led to inaccuracies and distortions. Here, we present a “roadmap” algorithm using spherical coordinates that allows the accurate localization of ligands bound to icosahedral or asymmetric virus surfaces.
Results and Discussion
The technique
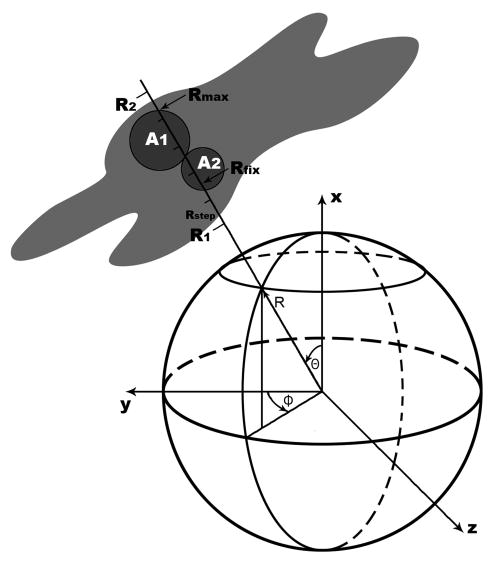
The RIVEM program was designed to project density within a radial shell surrounding a selected center onto the surface of a sphere. The positions of grid points within the input EM map are usually presented with respect to a Cartesian coordinate system (x, y, z), which can be redefined in terms of a spherical coordinate system (Θ, Φ, R) (Figure 1). The electron densities between the radii R1 and R2 can be sampled at Rstep intervals along a radius-vector (Θ, Φ). The density at each sampling step can be interpolated from the eight surrounding grid points in the original (x, y, z) map. The interpolated density values at the sampled points can be averaged and projected onto a sphere (Figure 1). The averaged density is then plotted onto a stereographic projection (Tong and Rossmann, 1997). The resultant plot can be contoured to show the position and height of the density relative to the orientation of the icosahedral axes (Figure 2).
Figure 1.
The spherical coordinate system and the plotting procedure in the RIVEM computer program. The relationship is shown between a Cartesian and spherical coordinate system as used in the program RIVEM. The electron density (gray area) between radii R1 and R2 or at a fixed radius (Rfix) can be projected onto the sphere. Two atoms A1 and A2 surrounded by their van der Waals radii are shown as filled circles. When a surface is plotted onto a spherical roadmap, atom A1, whose maximum distance from the origin (Rmax) is greater than that for atom A2, will be projected onto the sphere, thus identifying the atoms on the molecular surface. However, if a spherical section is being plotted at a radius Rfix, atom A2 (but not atom A1) would be chosen as its van der Waals sphere intersects the sphere with a radius of Rfix to the center of the virus.
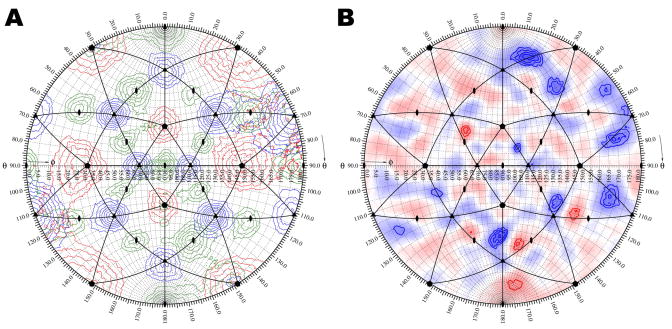
Figure 2.
Determination of the icosahedral axis positions in an asymmetric reconstruction of an icosahedral virus. A. Self-rotation function calculated for an asymmetric reconstruction of CPV. Rotation function peaks of the five-fold (kappa = 72°), three-fold (kappa = 120°), and two-fold (kappa = 180°) sections are shown as contours and colored red, blue, and green, respectively. The position of the symmetry axes for the mean orientation of an icosahedron is labeled with corresponding symbols. Icosahedral asymmetric units are outlined in black. B. Densities in the shell between 110 and 130 Å radii of a cryo-EM difference map projected onto a sphere and plotted as a stereographic projection. The map was of the difference between an asymmetrically reconstructed and icosahedrally reconstructed CPV capsid. Positive and negative densities are shown in blue and red, respectively. Contours are at intervals of one sigma above and below the mean value of the difference map.
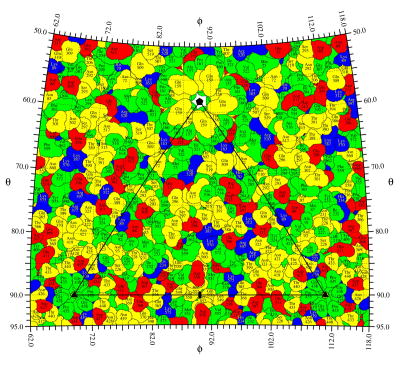
The same procedure can be used to plot the exposed surface area in terms of specific atoms or residues. Each atom is considered to be a sphere with a given van der Waals radius, RVDW. If desired, the atom's temperature factor, B, can be applied to extend the atom radius to Rexd, where Rexd = RVDW * [B/(8π2)]1/2. An additional overall temperature factor can be used to further increase the radii of the atomic spheres to simulate a low resolution cryo-EM map. The sphere around a single atom might be intercepted by several radius-vectors depending on the angular step intervals in Θ and Φ and the assumed atomic radius. Each of these vectors will intercept the sphere twice from which the larger distance from the center of the sphere (Rmax) is selected (Figure 1). The atom closest to the external surface along the specific radius-vector will then be the one with the largest Rmax (atom A1 in Figure 1) and will be chosen to represent the surface at this (Θ, Φ) position on the stereographic projection. Areas with atoms that belong to the same residue can be outlined by a border and associated with the residue name and number, resulting in a roadmap representation of the projected external surface (Figures 3 and 4). The angular intervals used to explore Θ and Φ can be sufficiently small (a useful increment is about 0.1°) to allow a good representation of the actual exposed area of each residue (Figures 3 and 4) in contrast to the fixed square unit area used by the earlier roadmap programs (Chapman, 1993; Rossmann and Palmenberg, 1988). By superimposing the projected density contours onto the roadmap of surface residues, the positional relationship between the amino acid residues and the density can be accurately interpreted (Figure 4C).
Figure 3.
A spherical roadmap of CPV surface residues. Basic, acid, polar, and hydrophobic residue are colored blue, red, yellow, and green, respectively. A little more than one icosahedral asymmetric unit is shown. The borders of the asymmetric unit are outlined in black, and the icosahedral symmetry axes are labeled with corresponding symbols.
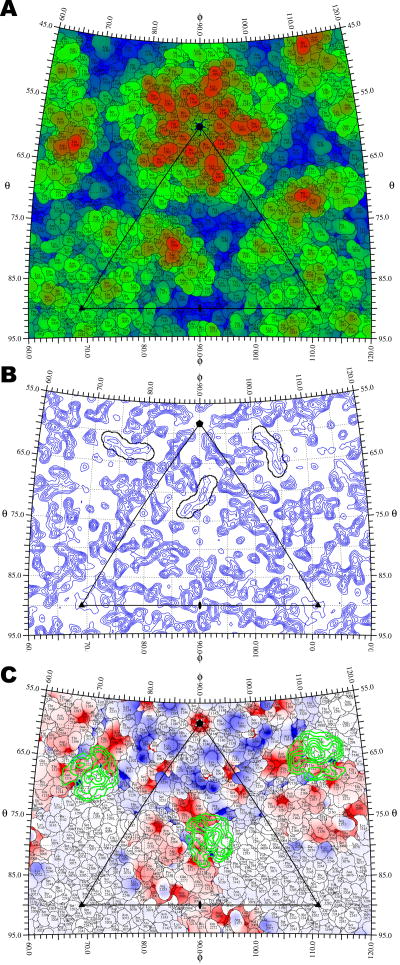
Figure 4.
Structure of CVA21 and its interaction with ICAM-1. A. Surface residues of CVA21 are plotted onto a stereographic projection and colored from blue (135 Å) to red (165 Å) based on their maximum radial distance from the center of the virus. B. The location of the “pocket factor” in a 3.2 Å resolution electron density map of CVA21 crystal (Xiao et al., 2005a) shown as a spherical section at a fixed radius (Rfix = 129.6 Å) is outlined in black based on the coordinates of a myristate molecule that was fitted to the density. C. The footprint of ICAM-1 onto the CVA21 surface. The difference density between 145 Å and 160 Å radii, isolating the ICAM-1 receptor, is projected onto a stereographic diagram and contoured in green at 1.5 sigma intervals above the mean density. The roadmap is colored according to the charge potential of CVA21 calculated by the program Delphi (Gilson and Honig, 1988).
Sometimes it is useful to plot the density on the surface of a defined sphere (radius Rfix), as opposed to projecting a shell of density. In this case, it is more appropriate to plot only the atoms that are within their van der Waals distance of the surface within radius Rfix (Figure 4B). In addition, the projected density can also be of a polygonal instead of spherical shell, which is useful when the shape of the virion or its membrane is an icosahedron (Böttcher et al., 1997a; Yan et al., 2000).
Various coloring schemes can be used to represent an assortment of features on spherical roadmaps. Specific residues, such as those studied by mutagenesis, can be colored to emphasize their position relative to a bound ligand. The atomic distance from the center of the virus can be used as a coloring scale to show the surface topology (Figure 4A). Other coloring schemes can be used to represent the height of projected density (Figure 2B), amino acid types (Figure 3), or electrostatic potential (Figure 4C).
Information about the orientation of symmetry elements (icosahedral operators for many viruses, five-fold symmetry for phage heads, and so forth) that can be used, for instance, to define the limits of an asymmetric unit is often helpful for the interpretation of EM maps. Furthermore, it is necessary to use the symmetry information to generate all symmetry related atoms before determining surface residues. Another use of the symmetry information is to impose averaging between equivalent density features.
Although the program RIVEM was initially developed for studying asymmetric features on icosahedral spherical viruses, it can also be used to investigate symmetry mismatched features in bacteriophages and other molecular complexes. Currently, the program supports only the X-Plor map format (Brünger et al., 1998), but other map formats can be added easily. Various plotting options combined with appropriate symmetry operators allow RIVEM to be used for globular protein studies, such as plots of electrostatic potential maps (Baker et al., 2001; Gilson and Honig, 1988) (Figure 4C) for each of the two surfaces of a protein-protein interaction.
Application to parvoviruses
The structure of canine parvovirus (CPV) was analyzed by means of an asymmetric cryo-EM reconstruction whose resolution had been estimated to be 30 Å (Hafenstein et al., 2006). The orientation of the viral capsid in the final asymmetric reconstruction was determined with a self-rotation function (Tong and Rossmann, 1997) using structure factors calculated by Fourier transformation of the cryo-EM density map (Figure 2A). Although no icosahedral symmetry had been applied during the cryo-EM reconstruction, dominant, icosahedrally distributed, rotation function peaks were found (Figure 2A). A difference map was then calculated between a cryo-EM reconstruction assuming icosahedral symmetry and an asymmetric reconstruction that had been reoriented to the same standard icosahedral axial system (Figure 2B). The difference density between 110 and 130 Å radii, corresponding to the protein shell, was plotted onto a stereographic projection using RIVEM (Hafenstein et al., 2006) (Figure 2B). This projection could be readily interpreted in terms of possible conformational changes relative to the symmetry axes, although the height of the differences were too low to establish the significance of the results at 30 Å resolution. A similar procedure had been used to investigate the icosahedral character of the heavy atom distribution in the analysis of Southern Bean Mosaic virus (Rayment et al., 1978).
Application to picornaviruses
The interaction between picornaviruses and their receptors has been studied by combining X-ray crystallography and cryo-EM image analysis (Rossmann et al., 2002). Many of the cellular receptors used by picornaviruses belong to the immunoglobulin superfamily and bind into a canyon-like depression on the viral surface surrounding each icosahedral five-fold vertex (Figure 4A). The difference density between the cryo-EM determined structure of the virus/receptor complex and the crystallographically determined virus structure calculated at the same resolution can be accurately projected onto a roadmap of the virus surface. One example given here is the projection of the density corresponding to the intercellular adhesion molecule-1 (ICAM-1) as seen projected onto the surface residues of the crystallographically determined coxsackievirus A21 (CVA21) structure (Xiao et al., 2005a) (Figure 4C). Another example is a plot of the density at a fixed radius Rfix to visualize the position of the “pocket factor” in CVA21 (Xiao et al., 2005a). This factor is a fatty acid-like molecule that is bound into a pocket immediately below the floor of the canyon, critical to the stability of the virion (Rossmann et al., 2002) (Figure 4B). The projected section provides an easy to interpret and accurate plot of the environment of the pocket factor within an icosahedral axial system.
Availability of program
The program RIVEM and its source code are freely available at http://structure.bio.purdue.edu/~viruswww/Rossmann_home/softwares.shtml.
Acknowledgments
We thank Jianghai Zhu, Ricardo Bernal, Victor Kostyuchenko, Wei Zhang, and Tao Sun for helpful discussions during the development of the RIVEM program, Susan Hafenstein for providing input data, and Sharon Wilder for help in the preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci US. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Belnap DM, McDermott BM, Jr, Filman DJ, Cheng N, Trus BL, Zuccola HJ, Racaniello VR, Hogle JM, Steven AC. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci US. 2000;97:73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal RA, Stock D. Three-dimensional structure of the intact Thermus thermophilus H+- ATPase/synthase by electron microscopy. Structure. 2004;12:1789–1798. doi: 10.1016/j.str.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Böttcher B, Kiselev NA, Stel'Mashchuk VY, Perevozchikova NA, Borisov AV, Crowther RA. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J Virol. 1997a;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature (London) 1997b;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bubeck D, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat Struct Mol Biol. 2005;12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli ME, Trus BL, Smith CS, Cheng N, Conway JF, Steven AC. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J Mol Biol. 2003;327:1–6. doi: 10.1016/s0022-2836(03)00117-7. [DOI] [PubMed] [Google Scholar]
- Chapman MS. Mapping the surface properties of macromolecules. Protein Science. 1993;2:459–469. doi: 10.1002/pro.5560020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ko Y, Delannoy M, Ludtke SJ, Chiu W, Pedersen PL. Mitochondrial ATP synthasome. Three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J Biol Chem. 2004;279:31761–31768. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Chiu W, McGough A, Sherman MB, Schmid MF. High-resolution electron cryomicroscopy of macromolecular assemblies. Trends Cell Biol. 1999;9:154–159. doi: 10.1016/s0962-8924(99)01511-1. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature (London) 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison SC, Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature London. 2004;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gillet A, Sanner M, Stoffler D, Olson A. Tangible interfaces for structural molecular biology. Structure. 2005;13:483–491. doi: 10.1016/j.str.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Gilson MK, Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J Mol Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Fuller SD, Stuart DI. Complementing crystallography: the role of cryo-electron microscopy in structural biology. Acta Crystallogr sect D. 1999;55:1742–1749. doi: 10.1107/s0907444999009956. [DOI] [PubMed] [Google Scholar]
- Hafenstein S, Palermo LM, Xiao C, Morais MC, Chipman PR, Bowman VD, Battisti AJ, Parrish CR, Rossmann MG. Asymmetric binding of transferrin receptor to parvovirus capsids. 2006 doi: 10.1073/pnas.0701574104. Manuscript in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, Blaas D. The cellular receptor of human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 2000;19:6317–6325. doi: 10.1093/emboj/19.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Maurizi MR, Steven AC. The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J Struct Biol. 2004;146:180–188. doi: 10.1016/j.jsb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature London. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, B GE, Prevelige PE, Jr, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006 doi: 10.1126/science.1127981. in press. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature London. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 Å resolution by single particle electron cryomicroscopy. Structure. 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Matadeen R, Patwardhan A, Gowen B, Orlova EV, Pape T, Cuff M, Mueller F, Brimacombe R, van Heel M. The Escherichia coli large ribosomal subunit at 7.5 Å resolution. Structure. 1999;7:1575–1583. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of φ29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-a visualization system for exploratory research and analysis. J Computat Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rayment I, Johnson JE, Suck D, Akimoto T, Rossmann MG. An 11Å resolution electron density map of southern bean mosaic virus. Acta Crystallogr sect B. 1978;34:567–578. [Google Scholar]
- Roseman AM. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr sect D. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Arisaka F, Battisti AJ, Bowman VD, Chipman PR, Fokine A, Hafenstein S, Kanamaru S, Kostyuchenko VA, Mesyanzhinov VV, Shneider MM, Morais MC, Leiman PG, Palermo LM, Parrish CR, Xiao C. From structure of the complex to understanding of the biology. Acta Crystallogr sect D. 2006 doi: 10.1107/S0907444906047330. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, He Y, Kuhn RJ. Picornavirus-receptor interactions. Trends Microbiol. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Palmenberg AC. Conservation of the putative receptor attachment site in picornaviruses. Virology. 1988;164:373–382. doi: 10.1016/0042-6822(88)90550-8. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. Structure of Ca2+ release channel at 14 Å resolution. J Mol Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Rossmann MG. Rotation function calculations with GLRF program. Meth Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- Van Etten JL, Lane LC, Meints RH. Viruses and viruslike particles of eukaryotic algae. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Quart Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Meth Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- Walz J, Koster AJ, Tamura T, Baumeister W. Capsids of tricorn protease studied by electron cryomicroscopy. J Struct Biol. 1999;128:65–68. doi: 10.1006/jsbi.1999.4169. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–189. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Xiao C, Bator-Kelly CM, Rieder E, Chipman PR, Craig A, Kuhn RJ, Wimmer E, Rossmann MG. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure. 2005a;13:1019–1033. doi: 10.1016/j.str.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005b;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- Yan X, Olson NH, Van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Building the atomic model for the bacterial flagellar filament by electron cryomicroscopy and image analysis. Structure. 2005;13:407–412. doi: 10.1016/j.str.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci US. 2001;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]