Abstract
Paradigms in drug design and discovery are changing at a significant pace. Concomitant to the sequencing of over 180 several genomes, the high-throughput miniaturization of chemical synthesis and biological evaluation of a multiple compounds on gene/protein expression and function opens the way to global drug-discovery approaches, no more focused on a single target but on an entire family of related proteins or on a full metabolic pathway. Chemogenomics is this emerging research field aimed at systematically studying the biological effect of a wide array of small molecular-weight ligands on a wide array of macromolecular targets. Since the quantity of existing data (compounds, targets and assays) and of produced information (gene/protein expression levels and binding constants) are too large for manual manipulation, information technologies play a crucial role in planning, analysing and predicting chemogenomic data. The present review will focus on predictive in silico chemogenomic approaches to foster rational drug design and derive information from the simultaneous biological evaluation of multiple compounds on multiple targets. State-of-the-art methods for navigating in either ligand or target space will be presented and concrete drug design applications will be mentioned.
Keywords: chemogenomics, ligand, target, virtual screening
Introduction
Until the recent sequencing of the human genome (Lander et al., 2001; Venter et al., 2001), drug discovery has long been a multidisciplinary effort to optimize ligands properties (potency, selectivity, pharmacokinetics) towards a single macromolecular target. It is estimated that, out of the 20–25 000 human genes supposed to encode for ca. 3000 druggable targets (Russ and Lampel, 2005), only a subset of that pharmacological space (ca. 800 proteins) has currently been investigated by the pharmaceutical industry (Paolini et al., 2006). Remarkably, medicinal chemistry followed a parallel boost with the miniaturization and parallelization of compound synthesis, such that over 10 million non-redundant chemical structures covers the actual chemical space, out of which ca. 1000 have been approved as drugs. Therefore, only a small fraction of compounds describing the current chemical space has been tested on a fraction of the entire target space. Chemogenomics is the new interdisciplinary field, which attempts to fully match target and ligand space, and ultimately identify all ligands of all targets (Caron et al., 2001). Various definitions of overlapping fields (chemical genetics, chemical genomics) have been proposed. We will herein consider a broad definition of chemogenomics encompassing chemoproteomics, namely the study of small-molecular-weight drug candidates on gene/protein function. From the definition of the field, one easily understands that chemogenomics will be at the interface of chemistry, biology and consequently informatics since data mining is required to extract reliable information. Furthermore, methodologies at the border of chemistry and biology (medicinal chemistry), chemistry and informatics (chemoinformatics), biology and informatics (bioinformatics) will also play a major role in bringing these major disciplines together. Chemogenomic approaches to drug discovery rely on at least three components, each necessitating hard experimental work: (1) a compound library, (2) a representative biological system (target library, single cell and whole organism), and (3) a reliable readout (for example, gene/protein expression, high-throughput binding or functional assay). By definition, analysing chemogenomic data is a never-ending learning process aimed at completing a two-dimensional (2-D) matrix, where targets/genes are usually reported as columns and compounds as rows, and where reported values are usually binding constants (Ki, IC50) or functional effects (for example, EC50). This matrix is sparse as far as all possible compounds have not been tested on all possible genes/proteins. Predictive chemogenomics will thus attempt to fill existing holes by predicting compounds–genes/proteins relationships. In silico approaches to predict such data (target selectivity for various ligands and ligand selectivity for various targets) will span pure ligand-based approaches (comparison of known ligands to predict their most probable targets), pure target-based approaches (comparison of targets or ligand-binding sites to predict their most likely ligands) or ultimately target-ligand based approaches (using experimental and predicted binding affinity matrices).
Description of ligand and target spaces
Basic assumptions of any chemogenomic-based approach are twofold: (i) compounds sharing some chemical similarity should also share targets and (ii) targets sharing similar ligands should share similar patterns (binding sites). Filling the full theoretical chemogenomic matrix thus implies that data on ‘unliganded' targets should be gathered from the closest ‘liganded' neighbouring targets, and that data on ‘untargeted' ligands should be gathered from the closest ‘targeted' ligands. The true question is how to measure distances between two ligands or two targets.
Ligand space
To efficiently navigate in ligand space, one first needs to describe the compound using appropriate properties (descriptors) and then to use a master equation to measure a distance between two compounds (similarity metric).
Descriptors are usually classified according to their dimensionality ranging from one dimensional (1-D) to three-dimensional (3-D) properties (Bender and Glen, 2004) (Figure 1 and Table 1) 1-D descriptors are easy and fast to compute. They describe global properties (for example, molecular weight, atom and bond counts), which can be derived from the chemical formulae and which are used in combination to predict absorption, distribution, metabolism, excretion and toxicity properties such as aqueous solubility (Votano et al., 2004), 1-octanol-water partition coefficient (Clark, 2005), plasma protein binding or bioavailability (Wang et al., 2006), but also to classify compounds (for example, drugs vs nondrugs (Sadowski and Kubinyi, 1998)) or ligands from various target families (Morphy, 2006) by linear or non-linear quantitative structure–activity relationships/quantitative structure–property relationships (QSAR/QSPR) methods. To fasten comparisons, 1-D linear representations of compounds are often used. The most popular of this kind of simplified string is the ‘Simplified Molecular Input Line Entry System' or SMILES (Weininger, 1988).
Figure 1.
Examples of molecular descriptors for small-molecular-weight ligands
Table 1.
Ligand descriptors
| Dimension | Nature | Examples |
|---|---|---|
| 1-D | Global | Molecular weight, atom and bound counts (for example, number of H-bond donors, number of rings), polar surface area, polarizability, log P) |
| 2-D | Topological | Topological and connectivity indices, fragments, substructures (for example, maximum common substructures), topological fingerprints (for example, structural keys) |
| 3-D | Conformational | n-points pharmacophore, shape, field, spectra and fingerprints |
Most ligand descriptors range in the family of 2-D topological descriptors, where the connectivity table (list of atoms and bonds) is parsed to encode both atomic and bond properties. The most intuitive way to represent this kind of information is the 2-D sketch of the structure (Figure 1), which enables to browse a ligand library for compounds sharing a particular 2-D motif (fragment, substructure). Graph-based methods which transforms the 2-D structure into a molecular graph (atoms being the nodes) are relatively popular for substructure search and clustering chemical compounds into subfamilies (Raymond et al., 2003), but present the noticeable disadvantage to be computationally slow. Much faster are fingerprint-based methods (Willett, 2006), where the occurrence of predefined structural events (atoms, fragments, rings, substructures and 2-D pharmacophores) are encoded into bit strings (sequence of ‘0' and ‘1' digits) called ‘fingerprints' which ere easy to derive, handle and compare. Although receptor-ligand recognition is a 3-D event, 2-D fingerprints have been found repeatedly more appropriate true 3-D fingerprints for similarity searches (Sheridan and Kearsley, 2002). Latter descriptors encode conformation-specific properties (atomic coordinates, 3-D pharmacophores, shapes, potentials, fields, spectra; Table 1), and therefore usually necessitate a common alignment of molecules to be compared in the same 3-D Cartesian space (especially if grid-based fields or potentials have to be compared) and a relevant sampling of conformational space accessible to each ligand. To avoid the alignment step which may cause false positives in a virtual screen, 3-D information can be translated into a bit string, which stores the occurrence of all possible pharmacophore tuplets (doublets, triplets and quadruplets) with their corresponding features (for example, H-bond acceptor, positively ionisable atom, and so on) and interfeature distances. Hence, comparing bit strings is much easier than comparing structures. Most similarity searches prefer a binary representation of 2- or 3-D properties to derive simple similarity indices, the most popular being the Tanimoto coefficient (Equation 1)
 |
a is the count of bits on in compound A, b is the count of bits on in compound B and c is the count of the bits on in both compound A and B.
The Tanimoto coefficient will thus range from 0 for two completely dissimilar structures to 1 for two identical compounds.
Target space
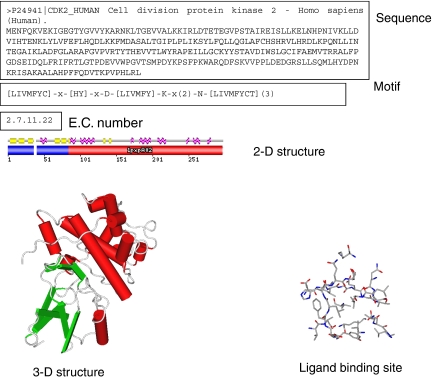
Proteins are commonly classified according to their sequence and structure (Table 2). The full amino-acid sequence is the very first interesting information (Figure 2), which already enables a reliable clustering of targets by family (for example, G protein-coupled receptors (GPCRs) and kinases). However, sequence lengths may considerably vary within a protein family (for example, sequence lengths of human GPCRs range from 290 to 6200 residues), such that analysing similarities and differences first requires an alignment of amino-acid sequences which can be tricky in case of large insertions/deletions. Therefore, one may focus on specific motifs (Attwood et al., 2003), which are a collection of continuous residues specific of a protein family (for example, DRY motif in TM III of rhodopsin-like GPCRs). To take into account the structural organization of the target, it can be of interest to look at the 2-D structure (mapping of α-helices, β-sheets, coils and random structures) and even better at the 3-D structure (atomic coordinates provided by X-ray diffraction, NMR or molecular modelling) and/or the corresponding fold. In chemogenomics-related approaches, one usually focuses on the ligand-binding site, where structural similarities among related targets are usually much higher than when considering the full 1-D sequence or 3-D structure.
Table 2.
Structural classification of proteins
| Dimension | Classification scheme | Databases |
|---|---|---|
| 1-D | Sequence | UniProt (Wu et al., 2006) and Pfam (Finn et al., 2006) |
| Patterns | PRINTS (Attwood et al., 2003) and PROSITE (Hulo et al., 2006) | |
| 2-D | Secondary structure fold | SCOP (Casbon and Saqi, 2005) and CATH (Reeves et al., 2006) |
| 3-D | Atomic coordinates | PDB (Berman et al., 2000) and MODBASE (Pieper et al., 2006) |
| binding site | Binding MOAD (Hu et al., 2005) and sc-PDB (Kellenberger et al., 2006) |
Abbreviations: MOAD, Mother of All Databases; UniProt, The Universal Protein Resource.
Figure 2.
Various representations of a protein using 1-D to 3-D properties.
Targets may also be classified according to their pharmacological profile (binding affinity for a panel of ligands) which means according to the nature of ligands they recognize (Paolini et al., 2006). Of course, there is a considerable overlap between sequence- and ligand-based classifications, since ligands generally bind to a subset of the protein universe. However, relationships across protein subfamilies are particularly interesting in drug design for predicting/modifying the pharmacological profile of a drug.
Target–ligand space
It is possible to directly navigate in the protein–ligand space by browsing full matrices in which either affinity or structural information is stored. Experimental evaluation of x compounds on y targets (for example, in vitro binding affinity assay) leads to a matrix of xy numbers (for example, IC50 values), which can be used to predict the affinity of a new compound to an existing target by multivariate linear regression (Kauvar et al., 1995), measure a structure–activity relationships distance between two targets (Vieth et al., 2004) and predict a global pharmacological profile (Krejsa et al., 2003). A clear advantage of this approach is that it relies on true binding affinity values and that experimentally derived descriptors will usually outperform computed descriptors. A clear drawback is the enormous amount of data required to derive true information such that similar approaches are not realistic, for example, in an academic environment. Therefore, one might substitute experimental with predicted affinities derived from either docking or 3-D QSAR approaches (Matter and Schwab, 1999; Fukunishi et al., 2006), although extrapolation will be limited here in a tiny protein space. Since binding free energy is extremely difficult to predict, replacing affinity by molecular interaction descriptors is possible. Of particular interest are structural interaction fingerprints (IFPs) (Singh et al., 2006), which converts atomic coordinates of a protein–ligand complex into a bit string featuring for each residue of a binding site, the type of molecular interactions (for example, H-bond, aromatic interaction, hydrophobic contact) developed by a co-crystallized or docked ligand. Comparing a series of complexes between n ligands and a single protein or between one ligand and n-related proteins is then performed as for ligands by computing distances between 1-D IFPs (Figure 3).
Figure 3.
(a) Deriving and (b) comparing protein–ligand complexes by molecular interaction fingerprints. ‘0' and ‘1' digits are replaced by colour-coded squares for the ease of comparison (blue, hydrophobic interactions; green, aromatic interactions; red, hydrogen bonds).
Ligand-based chemogenomic approaches
Annotating ligand libraries
The basic paradigm underlying ligand-based chemogenomic approaches is that molecules sharing enough similarity to existing biologically annotated ligands have enhanced probability to share the same biological profile (Figure 4). It is therefore very important to annotate chemical libraries with biological information (targets, in vitro affinity data and ADMET properties). Over recent years, there has been a huge effort mainly from small biotech companies to compile such data by an exhaustive survey of literature and patent data (Table 3). Since chemogenomic approaches usually focus on target families, most of these archives are related to the most pharmaceutically important target families (GPCRs, kinases, nuclear hormone receptors (NHRs), proteases and phosphodiesterases).
Figure 4.
Structure–activity relationship homology flowchart.
Table 3.
Biologically annotated compound libraries
| Database | Description | Website |
|---|---|---|
| AurSCOPE | Target family-oriented knowledge database containing pharmacological and pharmacokinetical data for 160 000 GPCR ligands and 77 000 kinase inhibitors | http://www.aureus-pharma.com |
| Bioprint | Biological profile (in vitro and clinical data) of 2400 small-molecular-weight drugs and drug-like compounds | http://www.cerep.fr/ |
| ChemBank | Storage of 50 000 compounds and related biological properties in 441 high-throughput screening and small molecule microarray assays | http://chembank.broad.harvard.edu/ |
| ChemBioBase | Target centric ligand databases (GPCRs, kinases, PDE) | http://www.jubilantbiosys.com/ |
| Kinase knowledge base | kinase structure–activity and chemical synthesis data | http://www.eidogen-sertanty.com/ |
| MDL Drug Data Report | 132 000 biologically relevant compounds and well-defined derivatives | http://www.mdli.com/ |
| MedChem database | 650 000 compounds with biological and pharmacological information | http://www.gvkbio.com |
| StARLITe | Highly curated target-compound SAR relationships | http://www.inpharmatica.co.uk/ |
| Wombat | 154 236 entries over 307 700 biological activities on 1320 unique targets | http://sunsetmolecular.com/ |
A good example has been provided by Novartis scientists (Schuffenhauer et al., 2003) who linked chemical space to target space by merging fields from separate chemical and biological databases to provide a unified and searchable chemogenomic database. On one hand, over 110 000 pharmaceutical ligands were gathered from the MDL Drug Data Report (Table 3). On the other hand, annotation of targets was based on existing classifications for enzymes and receptors. Linking MDDR ‘activity keys' to the target classification scheme enabled the annotation of 53 000 compounds totalling 799 different activity keys and related targets. Since the target's sequence is linkable to the ligand, sequence-based similarity searches of ligands for protein homologues of liganded targets are therefore feasible. Annotated reference ligands for a particular GPCR were used as starting points to recover either new receptor ligands or ligands of receptors close to the reference GPCR. Interestingly, the efficiency of the virtual screening approach was dependent on the phylogenetic distance between the reference and the query targets. Another straightforward application of biologically annotated compound libraries is the design of target-directed combinatorial libraries (Savchuk et al., 2004) focusing on chemotypes preferred by a family of targets.
Natural products also cover a very interesting chemical space of biological relevance because of the evolutionary pressure put on these compounds to bind, usually through highly specific mechanisms, to particular targets. The chemical space spanned by biologically annotated natural products was described recently as a structural and hierarchical scaffold tree (Koch et al., 2005), which can be browsed to design natural product-oriented chemical libraries.
Biologically annotated compound libraries are a direct source of potentially new biological mechanisms to correct a phenotype. Root et al. (2003) designed a library of 2036 biologically active compounds covering 169 different biochemical mechanisms, which was shown to be structurally diverse and able to provide 85 hits in a cell viability and proliferation assay. Among the 85 hits, 27 were supposed to be active by new biochemical mechanisms.
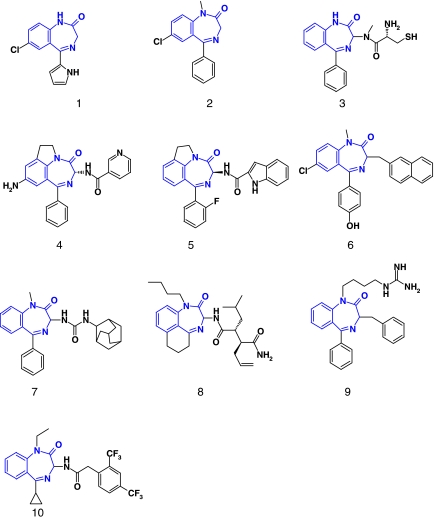
Privileged structures
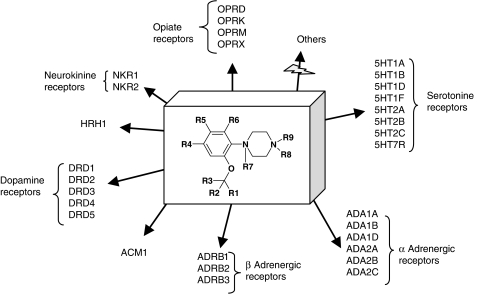
The term ‘privileged structure' was first coined by Evans et al. (1988), who noticed the promiscuity of the 1,4-benzodiazepine scaffold for various targets (Figure 5). A privileged structure is defined as ‘a substructure/scaffold exhibiting strong preferences for a particular area of the target space (for example, GPCRs) and suitable to orient the design of targeted compound libraries' (Klabunde and Hessler, 2002). In fact, a recent and deeper analysis of drug-like ligands show that privilege only appears upon a certain level of chemical functionalization of the scaffold (Schnur et al., 2006). For example, the biphenyl substructure is not a privileged structure but a simple protein-binding motif, since it occurs in a wide array of protein ligands with no particular preference for a certain target family. However, extending the biphenyl motif to a 2-tetrazolo-biphenyl dramatically enhance the specificity of the latter substructure for GPCRs (Schnur et al., 2006). Remarkably, many substructures apparently have corresponding binding sites in unrelated target families (for example, GPCRs, kinases, ion channels, proteases, nuclear hormone receptors). Only a few of them (see an example in Figure 6) are really selective for a certain target family (Schnur et al., 2006). A main reason for this exquisite specificity is that specific binding sites for peculiar substructure have been conserved along the evolution of target subfamilies (Bondensgaard et al., 2004; Surgand et al., 2006). Family-specific privileged structures are of prime importance to design targeted libraries and enhance hit rates when a protein from the targeted family is screened experimentally. A nice application of designing targeted libraries was presented by Amgen (Xia et al., 2004). After training a machine-learning algorithm to distinguish true kinase inhibitors from non-kinase inhibitors, multiple chemotypes could be selected to design a kinase-targeted library, which yielded high enrichment in true inhibitors in subsequent kinase inhibition assays.
Figure 5.
Permissivity of the 3H-1,4-benzodiazepin-2-one scaffold (in gray) across various targets. 1: Ro-5–3335, HIV-1 Tat inhibitor; 2: Diazepam, GABA-A receptor ligand; 3: 231023: farnesyltransferase inhibitor; 4: CI-1044, phosphodiesterase 4 inhibitor; 5: pranazepide, cholescystokinine (CCK) receptor antagonist; 6: BZ-423, F1F0 ATPase inhibitor, 7:171644, oxytocin receptor antagonist, 8: 309060: β-γ secretase inhibitor; 9: 278588: Stat5 agonist; 10: 276345: KVs channel blocker.
Figure 6.
Human GPCRs targeted by the orthoalkoxy-N-phenylpiperazine privileged structure (http://bioinfo-pharma.u-strasbg.fr/hGPCRLig/). Remarkably, no other proteins ever co-crystallized with drug-like compounds are able to recognize this substructure (http://bioinfo-pharma.u-strasbg.fr/scPDB/).
Ligand-based in silico screening
Main target families can be distinguished by a simple look at physicochemical properties (molecular weight, log P, polar surface area, H-bond donor and acceptor counts) of their cognate ligands (Morphy, 2006). One can thus easily imagine that more sophisticated descriptors can be used to predict a global target profile for any given compound, provided that targets to be predicted are sufficiently well described by existing ligands. Ligand-based in silico approaches to target fishing begin to appear in the literature (Cases et al., 2005; Bender et al., 2006; Bhavani et al., 2006; Mestres et al., 2006; Nettles et al., 2006; Nidhi et al., 2006; Steindl et al., 2006). They all share three basic components: (i) a set of reference compounds from which 2-D (scaffold, substructure, fingerprints) or 3-D descriptors (pharmacophore) are stored in a database, (ii) a screening procedure using either QSAR, machine learning (Bayesian classification, support vector machines) or pharmacophore searches and (iii) a screening collection to identify using above-described descriptors new molecules likely to share the same target or target profile than reference compounds (Figure 7).
Figure 7.
In silico target fishing approaches.
Mestres et al. (Cases et al., 2005; Mestres et al., 2006) have annotated a library of molecules targeting NHRs. Using a hierarchical classification for 2000 ligands and 25 receptors, chemogenomic links bridging ligand to target space can be easily recovered to distinguish selective from promiscuous scaffolds. Using Shannon Entropy descriptors (SHED) based on the distribution of atom-centred feature pairs, any compound collection can be screened to identify hits presenting SHED distances to a reference NHR ligand beyond a defined threshold and therefore likely to share the same NHR profile.
Novartis successfully applied a machine-learning algorithm using Bayesian statistics (Xia et al., 2004) to predict target profiles from extended connectivity fingerprints of compounds from the biologically annotated Wombat database (Nidhi et al., 2006). For each activity class (target), a separate Bayesian model is trained to distinguish known actives from known inactives. Predicting the most likely targets of compounds in the test set is then operated by predicting the probability of each test compound to be a ligand of each of the targets. On average, the correct target was found 77% of the time when training with Wombat compounds and testing molecules from another dataset (MDDR) over 10 different activity classes (Nidhi et al., 2006). A significant improvement in the predictions is observed when considering, instead of a series of individual probabilities, the global profile of all training compounds in which all target-associated probabilities are concatenated into a ‘Bayes affinity fingerprint' (Bender et al., 2006). Other 2- and 3-D descriptors have been assessed for the same application. 2-D descriptors were found to be more predictive with regard to correct target prediction than a pure 3-D pharmacophoric approach for test compounds structurally similar to those in the training set. For singletons (compounds exhibiting no strong similarity to molecules of the training set), the 3-D descriptor is more predictive.
In all these approaches, one must first automatically categorize compounds from the training set according to their molecular target without checking whether each compound really bind to its target, where (which binding site) and how (agonist or antagonist for receptor ligands). There is therefore a risk to train a machine-learning algorithm with incorrect data and to generate false rules. To overcome this drawback, more accurate but slower 3-D strategies are possible. Among them, a promising approach is to derive 3-D pharmacophores from protein–ligand complexes for which experimentally determined atomic coordinates and pharmacological activities exist (Steindl et al., 2006). The target-annotated pharmacophore database can be browsed to identify target(s) of new compounds by a classical pharmacophore search. The advantage of the method relies on the higher quality of the reference dataset, but is nevertheless limited by the pharmacophore generation step and the still limited chemical diversity observed among protein data bank (PDB) ligands (Kellenberger et al., 2006). For example, membrane receptors (for example, GPCRs, ion channels) cannot be predicted by this approach, crystallographic data being very sparse for these protein families, although homology model-based pharmacophores may be theoretically derived.
Target-based chemogenomic approaches
Controlling the selectivity of ligands towards related targets from the same family is crucial information in early drug-discovery stages. There is therefore a growing interest in comparing all targets from the same family especially those for which there is enough structural data (X-ray or NMR structures) to enable a proteome-wide comparative modelling of targets of still unknown structure (for example, protein kinases). Target-based chemogenomic approaches can be classified in two categories depending on whether the amino-acid sequence or the 3-D structure of targets is compared.
Sequence-based comparisons
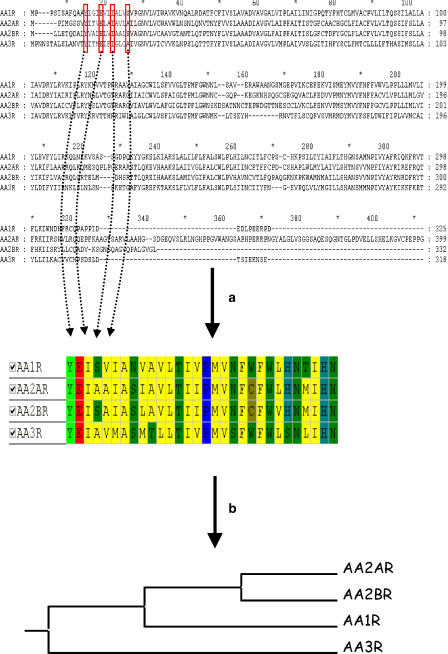
Sequence-based approaches are intended to be used for any kind of target family, provided that a multiple alignment of all targets to compare is reachable. They are generally used for target families where a lack of high-resolution structural data hampers target comparison. GPCRs constitute an ideal framework for sequence-based comparisons (Crossley, 2004; Frimurer et al., 2005; Kratochwil et al., 2005; Surgand et al., 2006), because it is a very important target family for drug design and only one member of this family (bovine rhodopsin) has been crystallized to date (Palczewski et al., 2000). After aligning all sequences, key residues supposed to map the binding site of most non-peptide ligands can be extracted and concatenated into an ungapped sequence of a few residues (Figure 8), which can be later used to derive a distance matrix based on sequence identity (Surgand et al., 2006), sequence similarity (Kratochwil et al., 2005) or physicochemical properties (Frimurer et al., 2005). An exhaustive cavity-based clustering of 372 human GPCRs has recently been proposed using such a strategy (Surgand et al., 2006). Interestingly, it reproduces perfectly the full sequence-based tree suggesting that only a few residues are really important when comparing targets across a family. This simplification enables a much simpler analysis of features (binding site regions), which are responsible for selective or permissive ligand binding by simply looking at residue conservation (Crossley, 2004; Surgand et al., 2006). There are several potential applications of cavity-based trees in drug discovery. A simple one consists in target hopping, which means discovering receptor ligands for a particular receptor by considering first the known ligands of closely related receptors. For example, CRTH2 receptor antagonists could have been identified from existing angiotensin II type 1 receptor antagonists (Frimurer et al., 2005), because both receptors were found close in the GPCR cavity-biased tree. In addition, the design of targeted libraries towards a particular area of the tree is facilitated by addressing those residues responsible for selectivity/promiscuity (Frimurer et al., 2005; Kratochwil et al., 2005).
Figure 8.
Sequence-based comparison of targets exemplified by human adenosine receptors (Surgand et al., 2006). (a) Selection of key cavity-lining residues and (b) Clustering according to residue conservation.
Structure-based comparisons
Structure-based comparisons are only possible for target families where there are enough good structural templates (X-ray structures) to afford the homology modelling of other related targets. In general, only ligand-binding sites (Hu et al., 2005; Kellenberger et al., 2006) are compared, since the basic aim of such comparisons is to understand the selectivity/permissivity features of related targets of known ligands.
A first possible strategy is to compare computed molecular interaction fields from the cavities to compare (Naumann and Matter, 2002; Hoppe et al., 2006; Pirard and Matter, 2006). Starting from a structural alignment of all targets, interaction energies generated by rolling several probe atoms (for example, sp3 carbon atom) at each point of 3-D grid encompassing the ligand-binding site are then concatenated into a MIF vector, which can be placed in a global matrix where rows describe targets and columns interaction energies at a given 3-D grid point (Figure 9) Comparing the MIFs and clustering the cognate targets can be done either by analysing the matrix by principal component analysis (Naumann and Matter, 2002; Pirard and Matter, 2006) or by calculating a MIF distance, which is later transformed in a target tree (Hoppe et al., 2006). A clear issue with this approach is that the comparison is highly dependent on the structural alignment, the grid resolution and the choice of the probe atoms. Moreover, it cannot be applied to targets of different families. However, its has been successfully applied to protein kinases (Naumann and Matter, 2002; Hoppe et al., 2006), serine proteases (Hoppe et al., 2006), matrix metalloproteinases (Pirard and Matter, 2006) and nuclear hormone receptors (Hoppe et al., 2006) to pinpoint cavity regions or subpockets explaining either selective or promiscuous ligand binding, and thus to guide the design of compound libraries towards the desirable selectivity pattern.
Figure 9.
Molecular interaction field (MIF)-based clustering of targets.
To avoid the previously reported structural alignment bias, 3-D atomic protein coordinates can be directly compared to measure a distance between two targets. Global structural alignment methods (Shindyalov and Bourne, 1998; Holm and Park, 2000; Standley et al., 2005) usually count the number of structurally equivalent residues by comparing overlapping sequence fragments. Such methods, however, do not work very well for discontinuous sequences (active sites) and for proteins exhibiting different folds. A second approach is to identify pre-defined structural motifs or templates (for example, Ser–His–Asp catalytic triad in serine proteases) and align a query to a reference protein by matching templates (Artymiuk et al., 1994; Wallace et al., 1997). However, numerous proteins (for example, kinases, GPCRs, ion channels) may share a binding site for a unique ligand (ATP) without sharing any structural template similarity. Most recent approaches to generate structural alignment describe proteins by physicochemical properties at representative locations. Molecular surfaces can be easily discretized in either chemically labelled sparse points (Rosen et al., 1998) or graphs (Kinoshita and Nakamura, 2003) and therefore aligned to maximize surface overlap with any reference. A database of protein surfaces (eF-site) has successfully been browsed to predict the function of a hypothetical archaeon protein (MJ0226) by detection of a mononucleotide binding site (Kinoshita and Nakamura, 2003). Surface-based comparisons are, however, relatively slow and thus incompatible with proteome-wide comparisons. Recent and faster methods (Schmitt et al., 2002; Jambon et al., 2003; Shulman-Peleg et al., 2004; Powers et al., 2006; Gold and Jackson, 2006) have been developed over the last 5 years. They all have in common to represent an active site of interest by pseudocenters (dummy atoms located along or close to every side chain of interest) encoding physicochemical properties (H-bonding capacity, aromaticity, hydrophobicity, charge) of their cognate residues, pseudocenters being linked together by edges and thus defining a molecular graph. Alignment is operated by detection of maximal common subgraphs (clique detection) (Gardiner et al., 1997) or geometric hashing (Nussinov and Wolfson, 1991) from defined pseudocenters. Local similarity at ligand binding subpockets can thus be detected for proteins with totally different folds and catalytic activities. Predicted similar binding sites can even be linked together in a global network to better position a protein in the target space (Zhang and Grigorov, 2006).
A nice example of binding site similarities for distant proteins has been exemplified by Weber et al. (2004), who detected cross-reactivity of arylsulfonamide-based COX-2 inhibitors with human carbonic anhydrase (HCA) based on the similarity of COX-2 and HCA binding pockets. A problem with these matching techniques is that the computed similarity score (usually dependent on the number of atom/pseudocenter/triangle matches) is not always easy to interpret, notably for active sites of different dimensions, because large actives sites will have a tendency to present more matches than small ones even if the latter are more similar. Therefore, normalized distance metrics similar to those used for comparing ligands are needed. A promising approach is proposed by Surgand (2006), who discretizes an active site by a dimensionless 80-triangle sphere and projects, from cβ atoms of cavity-lining residues to the sphere centre, various topological and physicochemical descriptors. A distance between two active sites is thus simply computed by summing up the normalized differences in descriptor space between each triangle of the sphere. The method was able to recognize remote binding site similarities (Figure 10) between a GPCR (GPR30) and a NHR (estrogen receptor α) sharing 4-hydroxytamoxifen as high-affinity ligand (Revankar et al., 2005).
Figure 10.
Screening a non-redundant subset of 1060 binding sites form the sc-PDB target library (Kellenberger et al., 2006) for ligand binding sites similar to that of GPR30 for 4-hydroxy-tamoxifen. (a) Ranking sc-PDB entries by decreasing similarity (ranging from 1 to 0) to the GPR30 4-hydroxy-tamoxifen binding site (extrapolated from a consensus list of 30 residues delimiting a canonical non-peptide binding site for most GPCR ligands as proposed by Surgand et al. (2006). The 3ert sc-PDB entry (4-hydroxy-tamoxifen binding site in the estrogen receptor α) is ranked second among 1060 investigated binding site. (b) Predicted alignment of GPR30 (blue) to ER-α binding site (green) for 4-OH tamoxifen (white ball and sticks). Both binding sites present a water-accessible negatively charged residue and a buried hydrophobic region (similarity index of 0.79 according to the SiteAlign program (Surgand et al., 2006)).
The current speed of such comparisons enables the definition of all-against-all similarity matrices (Schmitt et al., 2002; Shulman-Peleg et al., 2004; Gold and Jackson, 2006), and opens the door to various applications: (i) functional analysis and classification of ligand binding sites, (ii) predicting potential ligands, and (iii) anticipating side effects caused by targeting a peculiar protein.
An alternative approach to compare ligand-binding sites is to evaluate the similarity of potential ligand binding envelopes for known X-ray structure of apo or holoproteins (An et al., 2005). A first draft of the human pocketome, a collection of all possible ligand binding envelopes for a set of 943 crystallized human proteins, has been proposed recently (An et al., 2005) and clustered by envelope similarity. Interestingly, the ligand envelope-based tree only partially matches alternative trees based on the amino-acid sequence of the target proteins or on bound-ligand similarities (An et al., 2005).
Another recently proposed approach to compare proteins of the same family is to look at packing defects (Fernandez et al., 2004) localized at the so-called ‘dehydrons' (backbone heavy atoms with unsatisfied H-bonding partners), which are good indicators of protein capacity to interact with potential ligands and can be predicted form the amino-acid sequence. Packing distances between 32 PDB-reported kinases were shown to be almost identical to the pharmacological distance between these kinases estimated from an experimental affinity matrix derived for 17 inhibitors (Fernandez and Maddipati, 2006) and to efficiently guide the structure-based design of selective inhibitors for various enzymes specifically designed to target packing defects (Fernandez, 2005).
Target–ligand based chemogenomic approaches
Chemical annotation of target binding sites
Numerous biologically annotated chemical libraries can be browsed (Table 3) to link chemical to target spaces and focus ligand-based design to target families (Bender et al., 2006; Nettles et al., 2006; Nidhi et al., 2006). However, as far as information about the binding site is missing, there is a potential risk to compare compounds sharing the same target but not the same binding site (for example, orthosteric and allosteric ligands). It is therefore important to rigorously annotate protein sequences and/or binding site by the chemotype of the ligands they can recognize. The SMID (Small Molecule Interaction Database) archive is an interesting initiative to annotate protein amino-acid sequences by domain-specific ligands (Snyder et al., 2006). A total of 6300 ligands covering 230 000 experimentally observed domain/small molecule interactions have been stored in a relational database, which can be browsed to predict the most likely ligand of proteins of unknown 3-D structures by comparison of their domains to known protein structures using a reverse position-specific basic local alignment search (BLAST) procedures (Feldman et al., 2006). Ligand-annotated binding sites from the PDB are annotated in several databases (Kellenberger et al., 2006), but only two of them (BindingMOAD, sc-PDB; Table 3) consider the ligand from a pharmacological point of view and are therefore of interest for chemogenomic approaches. Such databases can be used to prioritize either ligands or molecular scaffolds for designing targeted compound libraries covering a well-defined target space (Figure 11).
Figure 11.
Querying the sc-PDB chemogenomic database (http://bioinfo-pharma.u-strasbg.fr/scPDB) for rule-of-five compliant (Lipinski et al., 2001) small molecular weight fragments (MW <300, clogP <3, H-bond donor count <3, H-bond acceptor count <6) co-crystallized with protein kinases.
2-D searches
To browse and predict protein–ligand complexes, one needs to set up simple descriptors for both ligands and proteins from knowledge databases (Table 3) and concatenate them into a single protein–ligand description. The easiest way to encode this information is to start form experimental binding affinity matrices (Kauvar et al., 1995; Krejsa et al., 2003; Vieth et al., 2004) and to define appropriate QSAR/QSPR models to predict the affinity of new compounds for registered targets or the full virtual profile by general neighbourhood behaviour modelling (Krejsa et al., 2003). Another approach has recently been proposed for deorphanizing GPCRs in which a ligand fingerprint is merged to a sequence-based target fingerprint if a high-affinity complex (pKi>7) has been reported in the PDSP database (http://pdsp.med.unc.edu/). A machine-learning algorithm was trained from 5319 non-redundant known complexes and applied to a set of 1 911 415 virtual complexes (55 orphan receptors and 34 753 drug-like compounds from the NCI database) to predict the most likely associations (Bock and Gough, 2005). Out-of-sample validations (finding the receptors of a promiscuous ligand and the ligands of a single target) were in general agreement with literature data and some predictions still awaiting experimental validations have been made.
3-D searches
A straightforward way to predict putative targets of ligands is to dock each of the ligands of the compound library into each of the active site of the target library. This strategy has been validated by several groups and proved able to recover the known ligands of known targets and predict their off-targets and thus some potential side effects (Chen and Zhi, 2001; Paul et al., 2004). Up to now, there is a single successful target fishing application described in the literature utilizing a docking approach (Muller et al., 2006). Hence, inverse docking requires first a high-quality 3-D dataset of binding sites whose automated set-up is quite difficult, and second an accurate scoring function to properly rank targets. A problem is that energy-based scoring functions are not very good at quantifying very heterogeneous protein–ligand complexes by decreasing binding-free energies (Ferrara et al., 2004) and that alternative ways of scoring are requested for efficient target selection. Among the most promising methods is the computation of IFPs between a protein and its ligand. Practically, IFPs are simple bit strings that convert 3-D information about protein–ligand interactions into simple 1-D bit vector representations (Figure 3) that can be quickly compared by the use of traditional metrics (for example, Tanimoto coefficient, Euclidian distance). Usage of IFPs have shown several promising features: (i) enhancing the quality of pose prediction in docking experiments (Deng et al., 2004; Marcou and Rognan, 2006); (ii) clustering protein–ligand interactions for a panel of related inhibitors according to the diversity of their interaction with a target subfamily (Chuaqui et al., 2005; Marcou and Rognan, 2006); (iii) assisting target-biased library design (Deng et al., 2006).
However, docking-independent 3-D methods may also constitute an interesting approach to predict protein–ligand complexes. A significant problem is to encode protein and ligand properties with similar descriptors such that one partner can be retrieved by using the second one as a query. A promising solution is proposed with the CoLiBRI (Complementary Ligands Based on Receptor Information) method (Oloff et al., 2006) in which both ligand and active site atoms are described by a same vector of molecular descriptors derived from shape and electronic properties of isolated atoms. Therefore, it is possible to directly correlate chemical similarities between active site and their ligands by mapping patterns of active sites onto patterns of their complementary ligands. When applied to a test data set of 800 high-resolution PDB complexes, the complementary ligand was ranked among the top 1% of a large library in 90% of tested active sites. Accuracy dropped significantly for active sites very different from those in the test set but still usable as a prefiltering step for removing the most improbable ligands (Oloff et al., 2006).
Concluding remarks
Chemogenomic approaches to rational drug discovery have been exploding in the last years as high-throughput data (structure, binding affinity and functional effects) become available for both targets and ligands of pharmaceutical interest. Numerous ways to link those data have been proposed focusing on either ligand or target neighbourhood. A clear data organization and storage is necessary to foster such applications and begins to emerge for the most interesting target families (kinases, GPCRs and NHRs). In a near feature, an earlier and better control of ligand selectivity can be anticipated by using chemogenomic data. This does not mean that more selective ligands are going to be designed, but simply that the observed selectivity profile of the compound will be compatible with a therapeutical usage. In addition, novel genomic targets could be better addressed after locating them in the target space and exploiting the associated chemical information.
Abbreviations
- 1-D
one-dimensional
- 2-D
two-dimensional
- 3-D
three-dimensional
- ADMET
absorption, distribution, metabolism, excretion and toxicity
- GPCR
G protein-coupled receptor
- NHR
nuclear hormone receptor
- QSPR
quantitative structure–property relationships
- QSAR
quantitative structure–activity relationships
Conflict of interest
The author states no conflict of interest.
References
- An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- Artymiuk PJ, Poirrette AR, Grindley HM, Rice DW, Willett P. A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J Mol Biol. 1994;243:327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Org Biomol Chem. 2004;2:3204–3218. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- Bender A, Jenkins JL, Glick M, Deng Z, Nettles JH, Davies JW. ‘Bayes affinity fingerprints' improve retrieval rates in virtual screening and define orthogonal bioactivity space: when are multitarget drugs a feasible concept. J Chem In Model. 2006;46:2445–2456. doi: 10.1021/ci600197y. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavani S, Nagargadde A, Thawani A, Sridhar V, Chandra N. Substructure-based support vector machine classifiers for prediction of adverse effects in diverse classes of drugs. J Chem Inf Model. 2006;46:2478–2486. doi: 10.1021/ci060128l. [DOI] [PubMed] [Google Scholar]
- Bock JR, Gough DA. Virtual screen for ligands of orphan G protein-coupled receptors. J Chem Inf Model. 2005;45:1402–1414. doi: 10.1021/ci050006d. [DOI] [PubMed] [Google Scholar]
- Bondensgaard K, Ankersen M, Thogersen H, Hansen BS, Wulff BS, Bywater RP. Recognition of privileged structures by G-protein coupled receptors. J Med Chem. 2004;47:888–899. doi: 10.1021/jm0309452. [DOI] [PubMed] [Google Scholar]
- Caron PR, Mullican MD, Mashal RD, Wilson KP, Su MS, Murcko MA. Chemogenomic approaches to drug discovery. Curr Opin Chem Biol. 2001;5:464–470. doi: 10.1016/s1367-5931(00)00229-5. [DOI] [PubMed] [Google Scholar]
- Casbon J, Saqi MA. S4: structure-based sequence alignments of SCOP superfamilies. Nucleic Acids Res. 2005;33:D219–D222. doi: 10.1093/nar/gki043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases M, Garcia-Serna R, Hettne K, Weeber M, van der Lei J, Boyer S, et al. Chemical and biological profiling of an annotated compound library directed to the nuclear receptor family. Curr Top Med Chem. 2005;5:763–772. doi: 10.2174/1568026054637665. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Zhi DG. Ligand–protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins. 2001;43:217–226. doi: 10.1002/1097-0134(20010501)43:2<217::aid-prot1032>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Chuaqui C, Deng Z, Singh J. Interaction profiles of protein kinase-inhibitor complexes and their application to virtual screening. J Med Chem. 2005;48:121–133. doi: 10.1021/jm049312t. [DOI] [PubMed] [Google Scholar]
- Clark M. Generalized fragment-substructure based property prediction method. J Chem Inf Model. 2005;45:30–38. doi: 10.1021/ci049744c. [DOI] [PubMed] [Google Scholar]
- Crossley R. The design of screening libraries targeted at G-protein coupled receptors. Curr Top Med Chem. 2004;4:581–588. doi: 10.2174/1568026043451140. [DOI] [PubMed] [Google Scholar]
- Deng Z, Chuaqui C, Singh J. Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein–ligand binding interactions. J Med Chem. 2004;47:337–344. doi: 10.1021/jm030331x. [DOI] [PubMed] [Google Scholar]
- Deng Z, Chuaqui C, Singh J. Knowledge-based design of target-focused libraries using protein–ligand interaction constraints. J Med Chem. 2006;49:490–500. doi: 10.1021/jm050381x. [DOI] [PubMed] [Google Scholar]
- Evans BE, Rittle KE, Bock MG, DiPardo RM, Freidinger RM, Whitter WL, et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem. 1988;31:2235–2346. doi: 10.1021/jm00120a002. [DOI] [PubMed] [Google Scholar]
- Feldman HJ, Snyder KA, Ticoll A, Pintilie G, Hogue CW. A complete small molecule dataset from the protein data bank. FEBS Lett. 2006;580:1649–1653. doi: 10.1016/j.febslet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Fernandez A. Incomplete protein packing as a selectivity filter in drug design. Structure. 2005;13:1829–1836. doi: 10.1016/j.str.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Maddipati S. A priori inference of cross reactivity for drug-targeted kinases. J Med Chem. 2006;49:3092–3100. doi: 10.1021/jm060163j. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Rogale K, Scott R, Scheraga HA. Inhibitor design by wrapping packing defects in HIV-1 proteins. Proc Natl Acad Sci USA. 2004;101:11640–11645. doi: 10.1073/pnas.0404641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL., III Assessing scoring functions for protein–ligand interactions. J Med Chem. 2004;47:3032–3047. doi: 10.1021/jm030489h. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frimurer TM, Ulven T, Elling CE, Gerlach LO, Kostenis E, Hogberg T. A physicogenetic method to assign ligand-binding relationships between 7TM receptors. Bioorg Med Chem Lett. 2005;15:3707–3712. doi: 10.1016/j.bmcl.2005.05.102. [DOI] [PubMed] [Google Scholar]
- Fukunishi Y, Kubota S, Nakamura H. Noise reduction method for molecular interaction energy: application to in silico drug screening and in silico target protein screening. J Chem Inf Model. 2006;46:2071–2084. doi: 10.1021/ci060152z. [DOI] [PubMed] [Google Scholar]
- Gardiner EJ, Artymiuk PJ, Willett P. Clique-detection algorithms for matching three-dimensional molecular structures. J Mol Graph Model. 1997;15:245–253. doi: 10.1016/s1093-3263(97)00089-2. [DOI] [PubMed] [Google Scholar]
- Gold ND, Jackson RM. Fold independent structural comparisons of protein–ligand binding sites for exploring functional relationships. J Mol Biol. 2006;355:1112–1124. doi: 10.1016/j.jmb.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Hoppe C, Steinbeck C, Wohlfahrt G. Classification and comparison of ligand-binding sites derived from grid-mapped knowledge-based potentials. J Mol Graph Model. 2006;24:328–340. doi: 10.1016/j.jmgm.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother of All Databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambon M, Imberty A, Deleage G, Geourjon C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- Kauvar LM, Higgins DL, Villar HO, Sportsman JR, Engqvist-Goldstein A, Bukar R, et al. Predicting ligand binding to proteins by affinity fingerprinting. Chem Biol. 1995;2:107–118. doi: 10.1016/1074-5521(95)90283-x. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank. J Chem Inf Model. 2006;46:717–727. doi: 10.1021/ci050372x. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Koch MA, Schuffenhauer A, Scheck M, Wetzel S, Casaulta M, Odermatt A, et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP) Proc Natl Acad Sci USA. 2005;102:17272–17277. doi: 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil NA, Malherbe P, Lindemann L, Ebeling M, Hoener MC, Muhlemann A, et al. An automated system for the analysis of G protein-coupled receptor transmembrane binding pockets: alignment, receptor-based pharmacophores, and their application. J Chem Inf Model. 2005;45:1324–1336. doi: 10.1021/ci050221u. [DOI] [PubMed] [Google Scholar]
- Krejsa CM, Horvath D, Rogalski SL, Penzotti JE, Mao B, Barbosa F, et al. Predicting ADME properties and side effects: the BioPrint approach. Curr Opin Drug Discov Devel. 2003;6:470–480. [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Marcou G, Rognan D. Optimizing scaffold docking by use of molecular interaction fingerprints. J Chem Info Model. 2006;47:195–207. doi: 10.1021/ci600342e. [DOI] [PubMed] [Google Scholar]
- Matter H, Schwab W. Affinity and selectivity of matrix metalloproteinase inhibitors: a chemometrical study from the perspective of ligands and proteins. J Med Chem. 1999;42:4506–4523. doi: 10.1021/jm990250u. [DOI] [PubMed] [Google Scholar]
- Mestres J, Martin-Couce L, Gregori-Puigjane E, Cases M, Boyer S. Ligand-based approach to in silico pharmacology: nuclear receptor profiling. J Chem Inf Model. 2006;46:2725–2736. doi: 10.1021/ci600300k. [DOI] [PubMed] [Google Scholar]
- Morphy R. The influence of target family and functional activity on the physicochemical properties of pre-clinical compounds. J Med Chem. 2006;49:2969–2978. doi: 10.1021/jm0512185. [DOI] [PubMed] [Google Scholar]
- Muller P, Lena G, Boilard E, Bezzine S, Lambeau G, Guichard G, et al. In silico guided target identification of a scaffold-focused library: 1,3,5-triazepan-2,6-diones as novel phospholipase A2 inhibitors. J Med Chem. 2006;49:6768–6778. doi: 10.1021/jm0606589. [DOI] [PubMed] [Google Scholar]
- Naumann T, Matter H. Structural classification of protein kinases using 3D molecular interaction field analysis of their ligand binding sites: target family landscapes. J Med Chem. 2002;45:2366–2378. doi: 10.1021/jm011002c. [DOI] [PubMed] [Google Scholar]
- Nettles JH, Jenkins JL, Bender A, Davies JW, Glick M. Bridging chemical and biological space: target fishing using 2D and 3D molecular descriptors. J Med Chem. 2006;49:6802–6810. doi: 10.1021/jm060902w. [DOI] [PubMed] [Google Scholar]
- Nidhi, Glick M, Davies JW, Jenkins JL. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J Chem Inf Model. 2006;46:1124–1133. doi: 10.1021/ci060003g. [DOI] [PubMed] [Google Scholar]
- Nussinov R, Wolfson HJ. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc Natl Acad Sci USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oloff S, Zhang S, Sukumar N, Breneman C, Tropsha A. Chemometric analysis of ligand receptor complementarity: identifying Complementary Ligands Based on Receptor Information (CoLiBRI) J Chem Inf Model. 2006;46:844–851. doi: 10.1021/ci050065r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: a g protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- Paul N, Kellenberger E, Bret G, Muller P, Rognan D. Recovering the true targets of specific ligands by virtual screening of the protein data bank. Proteins. 2004;54:671–680. doi: 10.1002/prot.10625. [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirard B, Matter H. Matrix metalloproteinase target family landscape: a chemometrical approach to ligand selectivity based on protein binding site analysis. J Med Chem. 2006;49:51–69. doi: 10.1021/jm050363f. [DOI] [PubMed] [Google Scholar]
- Powers R, Copeland JC, Germer K, Mercier KA, Ramanathan V, Revesz P. Comparison of protein active site structures for functional annotation of proteins and drug design. Proteins. 2006;65:124–135. doi: 10.1002/prot.21092. [DOI] [PubMed] [Google Scholar]
- Raymond JW, Blankley CJ, Willett P. Comparison of chemical clustering methods using graph- and fingerprint-based similarity measures. J Mol Graph Model. 2003;21:421–433. doi: 10.1016/s1093-3263(02)00188-2. [DOI] [PubMed] [Google Scholar]
- Reeves GA, Dallman TJ, Redfern OC, Akpor A, Orengo CA. Structural diversity of domain superfamilies in the CATH database. J Mol Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–1630. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Rosen M, Lin SL, Wolfson H, Nussinov R. Molecular shape comparisons in searches for active sites and functional similarity. Protein Eng. 1998;11:263–277. doi: 10.1093/protein/11.4.263. [DOI] [PubMed] [Google Scholar]
- Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- Sadowski J, Kubinyi H. A scoring scheme for discriminating between drugs and nondrugs. J Med Chem. 1998;41:3325–3329. doi: 10.1021/jm9706776. [DOI] [PubMed] [Google Scholar]
- Savchuk NP, Balakin KV, Tkachenko SE. Exploring the chemogenomic knowledge space with annotated chemical libraries. Curr Opin Chem Biol. 2004;8:412–417. doi: 10.1016/j.cbpa.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J Mol Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- Schnur DM, Hermsmeier MA, Tebben AJ. Are target-family-privileged substructures truly privileged. J Med Chem. 2006;49:2000–2009. doi: 10.1021/jm0502900. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer A, Floersheim P, Acklin P, Jacoby E. Similarity metrics for ligands reflecting the similarity of the target proteins. J Chem Inf Comput Sci. 2003;43:391–405. doi: 10.1021/ci025569t. [DOI] [PubMed] [Google Scholar]
- Sheridan RP, Kearsley SK. Why do we need so many chemical similarity search methods. Drug Discov Today. 2002;7:903–911. doi: 10.1016/s1359-6446(02)02411-x. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Deng Z, Narale G, Chuaqui C. Structural interaction fingerprints: a new approach to organizing, mining, analyzing, and designing protein-small molecule complexes. Chem Biol Drug Des. 2006;67:5–12. doi: 10.1111/j.1747-0285.2005.00323.x. [DOI] [PubMed] [Google Scholar]
- Snyder KA, Feldman HJ, Dumontier M, Salama JJ, Hogue CW. Domain-based small molecule binding site annotation. BMC Bioinformatics. 2006;7:152. doi: 10.1186/1471-2105-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley DM, Toh H, Nakamura H. GASH: an improved algorithm for maximizing the number of equivalent residues between two protein structures. BMC Bioinformatics. 2005;6:221. doi: 10.1186/1471-2105-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindl TM, Schuster D, Laggner C, Langer T. Parallel screening: a novel concept in pharmacophore modeling and virtual screening. J Chem Inf Model. 2006;46:2146–2157. doi: 10.1021/ci6002043. [DOI] [PubMed] [Google Scholar]
- Surgand JS. Développement de nouvelles méthodes bioinformatiques pour l'étude des récepteurs couplés aux protéines G. Thèse de l'Université Louis Pasteur – Strasbourg I: France; 2006. [Google Scholar]
- Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Kinomics-structural biology and chemogenomics of kinase inhibitors and targets. Biochim Biophys Acta. 2004;1697:243–257. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Votano JR, Parham M, Hall LH, Kier LB, Hall LM. Prediction of aqueous solubility based on large datasets using several QSPR models utilizing topological structure representation. Chem Biodivers. 2004;1:1829–1841. doi: 10.1002/cbdv.200490137. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Borkakoti N, Thornton JM. TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Krudy G, Xie XQ, Wu C, Holland G. Genetic algorithm-optimized QSPR models for bioavailability, protein binding, and urinary excretion. J Chem Inf Model. 2006;46:2674–2683. doi: 10.1021/ci060087t. [DOI] [PubMed] [Google Scholar]
- Weber A, Casini A, Heine A, Kuhn D, Supuran CT, Scozzafava A, et al. Unexpected nanomolar inhibition of carbonic anhydrase by COX-2-selective celecoxib: new pharmacological opportunities due to related binding site recognition. J Med Chem. 2004;47:550–557. doi: 10.1021/jm030912m. [DOI] [PubMed] [Google Scholar]
- Weininger D. SMILES 1. Introduction and encoding rules. J Chem Inf Comput Sci. 1988;28:31–36. [Google Scholar]
- Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Maliski EG, Gallant P, Rogers D. Classification of kinase inhibitors using a Bayesian model. J Med Chem. 2004;47:4463–4470. doi: 10.1021/jm0303195. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Grigorov MG. Similarity networks of protein binding sites. Proteins. 2006;62:470–478. doi: 10.1002/prot.20752. [DOI] [PubMed] [Google Scholar]