Abstract
Knowledge gained from the revolutions in genomics and proteomics has helped to identify many of the key molecules involved in cellular signalling. Researchers, both in academia and in the pharmaceutical industry, now screen, at a sub-cellular level, where and when these proteins interact. Fluorescence imaging and molecular labelling combine to provide a powerful tool for real-time functional biochemistry with molecular resolution. However, they traditionally have been work-intensive, required trained personnel, and suffered from low through-put due to sample preparation, loading and handling. The need for speeding up microscopy is apparent from the tremendous complexity of cellular signalling pathways, the inherent biological variability, as well as the possibility that the same molecule plays different roles in different sub-cellular compartments. Research institutes and companies have teamed up to develop imaging cytometers of ever-increasing complexity. However, to truly go high-speed, sub-cellular imaging must free itself from the rigid framework of current microscopes.
Keywords: fluorescence microscopy, quantitative imaging, subcellular localization, pattern recognition, tissue microarrays, cellular diagnostics, protein activity
Early microscopy is above all a history of instrument development. Skilled lens grinding, improved mechanical stability and careful matching of lenses together led to the building of 17th-century telescopes and microscopes. Botanists, physicians and anatomists began to draw, discuss and publish their observations in luxurious book editions that increasingly attracted a growing scientifically interested public. The manufacture of less expensive microscopes during Victorian times allowed far more people to see the world of microscopic organisms in a drop of pond water or to observe the formerly unimagined intricate details of the structure of animals, plants and crystals. Exhibitions or ‘conversazione' were organized where many microscopes stood side by side to demonstrate the wonders of the microscopic world. Despite impressive progress throughout the 19th and early 20th century, it is surprising how little microscopy has changed. A look into many of today's imaging facilities stunningly resembles the 19th-century engraving in Figure 1. All the elements are already there: microscopists preparing samples, gazing through the eyepieces of bulky microscopes, capturing images of a fairly small number of cells and excitedly sharing images. Certainly, these early microscopes have little to rival the resolving power, sensitivity and variety of different contrast modes offered by modern research instruments. Digital imaging has revolutionized image acquisition, processing and data handling, and genetically encoded fluorescent proteins can address fluorescent labels with unprecedented precision, but – besides these impressive technological developments – the way in which microscopic images are acquired, shared and analysed today in most laboratories is a not very different from what scientists have been doing over a century.
Figure 1.
Early high-throughput microscopy. A scientific conversazione with microscopes provided to view microscopic objects. Such events attracted a large public. Engraving from Illustrated London News 28 April 1855. With permission, © The Shrewsbury Museum.
Not so in the future. In this issue of the British Journal of Pharmacology, Starkuviene and Pepperkok (2007) make a case for automated microscopy. Focussing on quantitative, high-content, high-throughput, cellular fluorescence imaging, the authors discuss the recent progress in, and the potential of, sub-cellular imaging for drug discovery. Acknowledging the impact that high-throughput genomics and proteomics had on pharmaceutical research, they now blow the horn for microscopy going automated.
Evolving the microscope to an automated imaging platform
Genomics, proteomics, biochemistry and molecular biology assays have brought large-scale screening and robotics into biomedical and pharmaceutical research. They have considerably accelerated both bench-top experimenting and industrial discovery through handling more samples in a shorter time, using standardized sample conditions and multiplexing experimental protocols. There should be no reason why fluorescence microscopes should resist this evolution towards less user interference, higher sample turnover and automated data mining.
Fluorescence imaging is certainly among the most sensitive available tools because this allows specific molecular events to be recorded through time and space, in situ. Thus, ‘multidimensional' here refers to the combination of two- (xy-), three- (xyz-, volume) spatial dimensions, four (plus time), five (plus spectral emission/excitation wavelength) information. Sometimes, different polarization angles, different contrasts are intercalated, giving rise to even higher-dimensional data sets. The current technologies for achieving such measurements are inherently complex, alignment sensitive, time consuming and sufficiently demanding to impose important constraints on the size, throughput and capacity of platform integration of the experimental set-up. Owing to to the high magnification, one, or – at most – a few cells are simultaneously viewed on an image. Thus, scaling up current microscopies to higher throughput typically requires the sequential imaging of small fields of view at high magnification.
Quantitative microscopy requires the extraction of quantitative information hidden within the dynamics of the genome, transcriptosome and proteome activity inside live cells and tissues. In the context of multidimensional imaging microscopy, most manufacturers of traditional microscopes have partially automated their microscope bodies by replacing formally hand-operated controls such as focusing wheels by motor-driven controls. Many routine research microscopes are readily upgraded to more sophisticated functions like autofocus, scanning stages and illumination and detection multiplexing. With digital camera-based image acquisition, these traditionally shaped microscopes are used in automated, high-throughput and/or high-content screening. Digital image acquisition and analysis has complemented this evolution on the software side and considerably contributed to fostering automated quantitative imaging (see Carpenter, 2007 for a recent commentary). Even standard packages like MetaMorph, Image J or open-source software tools (Gordon et al., 2006) offer complex acquisition and analysis protocols.
Yet, from there to ‘screening by imaging' isolated individual cells is still far away. In the age of ‘cytomics' (http://www.cytomics.info; Valet and Tarnok, 2004) or ‘tissomics' (http://www.tissomics.info; Kriete and Boyce, 2004), new concepts are needed to better integrate microscopic imaging into the continuum of sample and liquid flow as well as to handle the ever growing bioinformatics data stream.
Evolving the microscope body towards an automatic multiparametric platform must consider the full integration of probe-manipulation systems, climate control, fluidic systems and automated probe feeding systems, probably in a sterile or conditioned environment.
Placing of multi-well plates or microtitre plates onto the microscope stage is often impeded by the binocular tube. The same constraints apply to non-imaging readout of sensors (electrical, electrochemical, and so on.) with wire connections. Among the currently commercially available inverted microscopes, only TILL's IMIC has been developed to specifically meet the requirements of automated microscopy (Geisler et al., 2006). Based on a beam multiplexing, multilevel concept, it offers a scaleable and modular system that accommodates wide-field epifluorescence, total internal reflection fluorescence and scanning microscopies and can be fitted into screening workstations, for example, by adding a robot for slide and plate loading. Yet, being derived from a single-beam microscope, it maintains the need for sequential imaging and sample handling.
Evotec's OPERA, Cellomics' CELLWORX and related systems are inspired by the plate readers used in pharmaceutical research; they run 96-, 384- or 1536-well microtitre plates and come in a sealed bench-top package and are already set up for high-throughput screening, but ‘only' offer epifluorescence (plus deconvolution) or confocal contrast, respectively. Thus, these highly integrated systems provide much less flexibility than the relatively open environment of a conventional body with its different input and output ports.
Multiplexed imaging with multiple-beam microscopes
Confocal microscopy is used widely for three-dimensional (3-D) biological imaging, but can be too slow for many applications. A new class of microscope slide scanners is being developed that reduces scanning time and increases throughput by beam multiplexing. Many of these systems rely on continuous slide scanning and feeding, along with flashlamp or pulsed-laser excitation to avoid motion artefacts. Abandoning full confocality, sweeping a line focus across the specimen is an inexpensive alternative, but it compromizes resolution. Another conceivable approach for scanning a large field of view is by means of parallel laser scanning microscopy, that is, multiple-spot scanning (as in spinning-disk confocals or LaVision's TRIMSCOPE 64-spot multiplexer for scanning microscopes). The use of digital micromirror devices offers an alternative via large arrays of rapidly reconfigurable micromirrors (Botvinick et al., 2000). The same principle is readily extended to fast wavelength multiplexing of a white-light supercontinuum (McConnell et al., 2006), offering even more flexibility without moving parts.
Imaging flow cytometry
However, all these systems have in common one critical constraint and bottleneck for high-speed imaging that comes from the attachment of the sample to an optically transparent surface. Hence, moving from one cell to another requires the movement of the stage relative to the optical axis, either by stage or beam scanning. The severity of this limitation becomes apparent when one considers that for biomedical research, cell-based assay and cell-diagnostic applications, some of the most important targets are non-adherent cells, for example, stem cells, systemic cancer cells and lymphocytes.
Certain features like average brightness, protein coexpression (measured after transfection of multiple spectral variants of fluorescent proteins) or proximity (as probed by fluorescence resonance energy transfer) can be measured on a single-cell basis by flow cytometry, but the same individual cell could traditionally not be viewed over time, and some features (including morphology, structure and subcellular localization) are lost in flow cytometry. With flow rates of 10 000 cells/s, flow cytometry is about 100 times faster than imaging cytometry. Another obvious advantage of using cells in suspension is the ease of sample handling, which brings the advantages of microfluidics (Breslauer et al., 2006) and cell sorting to fluorescence imaging.
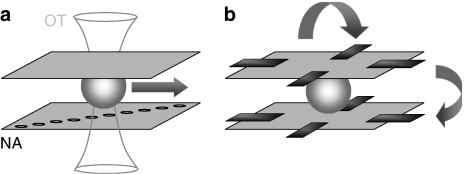
At present, microfluidics-based imaging still relies on bulky optical microscopes (10−2–10−1 m), although the microfluidic system itself is usually compact (10−4–10−5 m). Recently, the incorporation of optics into microfluidics has brought forth an interesting and novel microscopy geometry. Using an array of increasingly laterally displaced subwavelength (<200 nm) apertures oriented perpendicular to a linear flow, Heng et al. (2006a, 2006b) recently demonstrated near-diffraction limited imaging with a microfluidics-based lensless technique, termed ‘optofluidic microscopy' (OFM) (Figure 2a). In the present scheme, optical tweezers immobilize cells and then move them across the nanoaperture array with a constant speed. In this way, cells are able to remain in close proximity with the nanoaperture plane throughout scanning.
Figure 2.
Novel geometries for high-content, high-throughput microscopy. (a) Optofluidic microscopy (OFM). The major component of OFM is a metal-coated CMOS sensor array on which a linear array of subwavelength nanoapertures is patterned. This device is hermetically sealed on one side of a microfluidic delivery channel. The nanoaperture array is laid down in a slanted manner under microfluidic channel so that the illumination spot is scanned across the cell as the cell moves along with the fluid. Spatial resolution is defined by nanoaperture size and alleviates the constraints by the pixel width and pitch size of the underlying linear array charge-coupled device. OT, optical trap; NA, nanoaperture. (b) The cell rotator uses a DFC to trap and spatially position individual, non-adherent living cells suspended in low-conductance media. Periodic phase and intensity differences between the microelectrodes arranged in space to form a 3-D octode cage permit the generation of pN forces that rotate the cell around the focal plane so as to acquire a tomographic image. Combined with confocal detection, the method permits tomography-like multi-viewpoint 3-D imaging.
A different strategy providing multidimensional images with isotropic 3-D spatial resolution has been developed by Shorte and co-workers (Plate-Forme Imagerie Dynamique (PFID) at the Pasteur Institute) (Figure 2b). Non-adherent cells are supplied microfluidically and immobilized inside a small cage volume by an octagonal array of dielectrophoretic microelectrodes (DFCs). DFCs are fabricated photolithographically on transparent glass substrates and assembled face to face at ∼100–200-μm distance (Schnelle et al., 1993). Cells suspended in imaging buffer can thus be trapped and manipulated inside the cage. Alternating field strength and polarization creates pN forces (of the same order as in an optical trap), allowing to stably rotate the cell in the observation volume (Renaud et al., 2007). Hence, in place of acquiring a 3-D image by taking a z-stack along the optical axis, all imaging components are maintained in a fixed position and the cell is rotated around the focal plane. Snapshot images are taken at different rotation angles and rendered to give a tomographic image cube of the cell.
Future directions
Scaling up quantitative microscopy for high-content, high-throughput screening requires the seamless integration of advanced techniques coming from fields as diverse as optics, lithography, algorithm design, liquid handling and robotics. Individually, many of these fields are more advanced than the still emerging field of automated microscopy. For example, on data analysis, the extensive literature on pattern recognition and image segmentation has only sporadically made its way into the analysis of microscopic images (Boland et al., 1998; Zimmer et al., 2002), and, despite important progress in integrating motorized components into microscopes, the field is still far from the performance of modern robotics that has revolutionized the packaging or automobile industry.
One crucial question is: will automated microscopy remain the realm of large consortia and imaging platforms that combine transversal expertise in a tight and large-scale academic–industrial partnership, or will it, via commercial products, make its way into the small- and medium-sized laboratories to fundamentally change the way fluorescence imaging is done today in cell biological research? The ongoing evolution towards standardized cell culture, micropatterned substrates, automated liquid handling by microfluidics, along with integrated systems for automated digital image acquisition by robotic microscopy and quantitative analysis begins to offer the best of both worlds: microscopy is becoming simultaneously higher throughput and more quantitative at the single-cell level. Strangely, while much effort has been made in speeding up the individual steps of microscope-based cytometry, surprisingly little effort has gone into miniaturizing and streamlining the microscope core itself. Fluorescence excitation via planar optical waveguides, novel structured illumination schemes like dynamic speckle illumination that achieve quasi-confocality (Ventalon and Mertz, 2005) and the use of GRIN lenses and fibre optics instead of expensive objective lenses and rigid microscope frames will eventually combine to provide inexpensive imaging sensors that permit combinatorial microscopy (Axelrod, 2006) and not ‘just' multicolour confocal imaging. Progress in robotized microscopy will come from rethinking the light path of the microscope.
Acknowledgments
I thank JacSue Kehoe for critical reading of an earlier version of the paper and the Shrewsbury Museum Service for granting the reprint of Figure 1. Research related to automated microscopy in my lab is supported by the European Union (Grant nos. FP6-2004-013880, FP6-2005-019481, FP6-2006-037897).
Abbreviations
- DFC
dielectric field cage
- DMD
digital micromirror device
- DSI
dynamic speckle illumination
- FRET
fluorescence resonance energy transfer
- HTM
high-throughput microscopy
- OFM
optofluidic microscopy
- PLSM
parallel laser scanning microscopy
- TIRF
total internal reflection fluorescence
References
- Axelrod D. Combinatorial microscopy. Nat Rev Mol Cell Biol. 2006;7:944–952. doi: 10.1038/nrm2062. [DOI] [PubMed] [Google Scholar]
- Boland MV, Markey MK, Murphy RF. Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry. 1998;33:366–375. [PubMed] [Google Scholar]
- Botvinick EL, Li F, Cha S, Gough DA, Fainmain Y, Price JH.In vivo confocal microscopy based on the Texas Instruments digital micromirror device Optical Diagnostics of Living Cells III Proc SPIE 200012–20.In: Farkas DL, Leif R (eds).
- Breslauer DN, Lee PJ, Lee LP. Microfluidics-based systems biology. Mol BioSyst. 2006;2:97–112. doi: 10.1039/b515632g. [DOI] [PubMed] [Google Scholar]
- Carpenter AE. Software opens the door to quantitative imaging. Nat Methods. 2007;4:120–121. doi: 10.1038/nmeth0207-120. [DOI] [PubMed] [Google Scholar]
- Geisler T, Ressler JHH, Wolf B, Uhl R. Automated multiparametric platform for high-content and high-throughput analytical screening on living cells. IEEE Trans Rob Autom. 2006;3:169–176. [Google Scholar]
- Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods. 2006;4:175–181. doi: 10.1038/nmeth1008. [DOI] [PubMed] [Google Scholar]
- Heng X, Cui X, Knapp DW J, Yaqoob Z, McDowell EJ, Psaltis D, et al. Determining the resolution limit of nano aperture based optical imaging or sensing device. Opt Express. 2006a;14:10410–10425. doi: 10.1364/oe.14.010410. [DOI] [PubMed] [Google Scholar]
- Heng X, Erickson D, Baugh LR, Yaqoob Z, Sternberg PW, Psaltis D, et al. Optofluidic microscopy – a method for implementing a high-resolution optical microscope on a chip. Lab Chip. 2006b;6:1274–1276. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- Kriete A, Boyce K. Automated tissue analysis – a bioinformatics perspective. Methods Inf Med. 2004;44:32–37. [PubMed] [Google Scholar]
- McConnell G, Poland S, Girkin JM. Fast wavelength multiplexing of a white-light supercontinuum using a digital micromirror device for improved three-dimensional fluorescence microscopy. Rev Sci Instrum. 2006;77:013702. [Google Scholar]
- Renaud O, Heintzmann R, Saez-Cirion A, Schnelle T, Mueller T, Shorte S. A system and methodology for high-content visual screening of individual intact living cells in cell suspension. Proceedings of the SPIE. 2007. p. 64410Q.
- Schnelle T, Hagedorn R, Fuhr G, Fiedler S, Müller T. Three-dimensional electric field traps for manipulation of cells calculation and experimental verification. Biochim Biophys Acta. 1993;1157:127–140. doi: 10.1016/0304-4165(93)90056-e. [DOI] [PubMed] [Google Scholar]
- Starkuviene V, Pepperkok R.The potential of high-content high throughout microscopy in drug discovery Br J Pharmacol 200715262–71.(this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valet G, Leary JF, Tarnok A. Cytomics – new technologies: towards a human cytome project. Cytometry. 2004;59A:167–171. doi: 10.1002/cyto.a.20047. [DOI] [PubMed] [Google Scholar]
- Ventalon C, Mertz J. Quasi-confocal fluorescence sectioning with dynamic speckle illumination. Opt Lett. 2005;30:3350–3352. doi: 10.1364/ol.30.003350. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Labruyere E, Meas-Yedid V, Guillen N, Olivio-Marin JC. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: a tool for cell-based drug testing. IEEE Trans Med Imaging. 2002;21:1212–1221. doi: 10.1109/TMI.2002.806292. [DOI] [PubMed] [Google Scholar]