Abstract
Computational (in silico) methods have been developed and widely applied to pharmacology hypothesis development and testing. These in silico methods include databases, quantitative structure-activity relationships, similarity searching, pharmacophores, homology models and other molecular modeling, machine learning, data mining, network analysis tools and data analysis tools that use a computer. Such methods have seen frequent use in the discovery and optimization of novel molecules with affinity to a target, the clarification of absorption, distribution, metabolism, excretion and toxicity properties as well as physicochemical characterization. The first part of this review discussed the methods that have been used for virtual ligand and target-based screening and profiling to predict biological activity. The aim of this second part of the review is to illustrate some of the varied applications of in silico methods for pharmacology in terms of the targets addressed. We will also discuss some of the advantages and disadvantages of in silico methods with respect to in vitro and in vivo methods for pharmacology research. Our conclusion is that the in silico pharmacology paradigm is ongoing and presents a rich array of opportunities that will assist in expediating the discovery of new targets, and ultimately lead to compounds with predicted biological activity for these novel targets.
Keywords: quantitative structure–activity relationships, homology models, docking, pharmacology, ADME/Tox, in silico, in vitro, drug discovery, computational
Introduction
The first part of this review (Ekins et al., 2007) has briefly described the history and development of a field that can be globally referred to as in silico pharmacology. This included the development of methods and databases, quantitative structure–activity relationships (QSARs), similarity searching, pharmacophores, homology models and other molecular modelling, machine learning, data mining, network analysis and data analysis tools that all use a computer. We have also previously introduced how some of these methods can be used for virtual ligand- and target-based screening and virtual affinity profiling. In this second part of the review, we will greatly expand on the applications of these methods to many different target proteins and complex properties, and discuss the pharmacological space covered by some of these in silico efforts. In the process, we will detail the success of in silico methods at identifying new pharmacologically active molecules for many targets and highlight the resulting enrichment factors when screening active drug-like databases. We will finally discuss some of the advantages and disadvantages of in silico methods with respect to in vitro and in vivo methods for pharmacology research.
Pharmacological space covered
The applicability of computational approaches to ligand and target space in which a lead molecule against one gene family member is used for another similar target (termed chemogenomics) (Morphy et al., 2004; Sharom et al., 2004), will be discussed thoroughly in an upcoming review in this journal from Didier Rognan (personal communication) and will be only briefly addressed here. However, there have been several attempts to establish relationships between molecular structure and broad biological activity and effects that should be considered (see also section 2.3.1 in Ekins et al. (2007)) (Kauvar et al., 1995, 1998b; Kauvar and Laborde, 1998a). For example, the work of Fliri et al. (2005b) presented the biological spectra for a cross-section of the proteome. Using hierarchical clustering of the spectra similarity enabled a relationship between structure and bioactivity to be constructed. This work was extended to identify agonist and antagonist profiles at various receptors, correctly classifying similar functional activity in the absence of drug target information (Fliri et al., 2005c). Interestingly, using IC50 data as affinity fingerprints did not identify functional activity similarities between molecules as this approach was suggested to introduce a pharmacophoric bias (Fliri et al., 2005c). A similar probabilistic approach has also been applied by the same authors to link adverse effects for drugs (obtained from the drug labelling information) with biological spectra. For instance, clustering molecules by side effect profile showed that similar molecules had overlapping profiles, in the same way that they had similar biological spectra, linking preclinical with clinical effects (Fliri et al., 2005a). This work offers the intriguing possibility of predicting a biospectra profile, possible functional activity and a side effect profile for a new molecule based on similarity alone. However, confidence in this approach would be greatly enhanced by further prospective testing with a large test set of drug-like molecules not used to generate the underlying signature database.
A second group also from Pfizer presented a global mapping of pharmacological space and in particular focused on a polypharmacology network of molecules with activity against multiple proteins (Paolini et al., 2006). They have additionally generated Bayesian binary models (for molecules active at <10 μM or inactive) for 698 targets using over 200 000 molecules with biological data (from their in-house collection and the literature), suggesting that they would be useful for predicting primary pharmacology. Assessment of 617 approved oral drugs in two-dimensional (2D) molecular property space (molecular weight versus cLogP) showed that many of them had cLogP >5 and MW >500. In spite of this, their associated targets were potentially druggable but had yet to realize their potential (Paolini et al., 2006). Perhaps this work needs to be combined with that of Fliri and others for its true potential to be realized, to enable simultaneous understanding and prediction of target, proteomic, functional activity and side effects. A recent analysis using 48 molecular 2D descriptors followed by principal component (PCA) of over 12 000 anticancer molecules representing cancer medicinal chemistry space, showed that they populated a different space broader than hit-like space and orally available drug-like space. This would indicate that in order to find molecules for anticancer targets in commercially available databases, different rules are required other than those widely used for drug-likeness, as they may unfortunately filter out possible clinical candidates (Lloyd et al., 2006).
Methods to predict the potential biological targets for molecules from just chemical structure have been attempted by using different approaches to those already described above. For example, one study used probabilistic neural networks with 24 atom-type descriptors to classify 799 molecules from the MDL Drug Data Reports (MDDR) database with activity against one of the seven targets (G protein-coupled receptors (GPCRs), kinases, enzymes, nuclear hormone receptors and zinc peptidases) with excellent training, testing and prediction statistics (Niwa, 2004). Twenty-one targets related to depression were selected and molecules from the MDDR database were used to create support vector machine (SVM) classification models from atom-type descriptors (Lepp et al., 2006). These models had satisfactory predictions and recall values between 45 and 90%, the molecules recovered being on average of low molecular weight (<300) and some were active against more than one model. It was suggested that general SVM filters would be useful for virtual screening owing to their speed. Others have used similarity searching of the MDDR database against small numbers of reference inhibitors for several different targets and were able to show variable enrichment factors that were greater than random (Hert et al., 2004). The structure-based alternative to understanding small molecule–protein interactions is to flexibly dock molecules into multiple proteins. A representative of this inverse docking approach is INVDOCK, which was recently applied for identifying potential adverse reactions using a database of 147 proteins related to toxicities (DART). This method has been recently demonstrated with 11 marketed anti-HIV drugs resulting in reasonable accuracy against the DNA polymerase beta and DNA topoisomerase I (Ji et al., 2006).
The public availability of data on drugs and drug-like molecules may make the analyses described above possible for scientists outside the private sector. For example, chemical repositories such as DrugBank (http://redpoll.pharmacy.ualberta.ca/drugbank/) (Wishart et al., 2006), PubChem (http://pubchem.ncbi.nlm.nih.gov/), KiDB (http://kidb.bioc.cwru.edu/) (Roth et al., 2004; Strachan et al., 2006) and others consist of a wealth of target and small molecule data that can be mined and used for computational pharmacology approaches. Although much of the in silico pharmacology research to date has been focused on human targets, many of these databases contain data from other species that would also be useful for understanding species differences and promoting discovery of molecules for animal healthcare as well as assisting in understanding the significance of toxicological findings for chemicals released into the environment.
Examples of in silico pharmacology
To exhaustively describe all of the proteins that have been computationally modelled under the auspices of in silico pharmacology would be impossible in the confines of this review. Therefore, we will briefly overview the types of proteins that have been modelled and the methods used (see below and Table 1). In addition, we will focus on and describe particular pharmacological applications with regard to virtual screening where novel ligands have been identified. The reader is highly encouraged to study an extensive review of success stories in computer-aided design, which covers a large number of proteins that have been targets for all manner of in silico methods (Kubinyi, 2006), as well as other reviews that have dealt with the successes of individual methods (Fujita, 1997; Kurogi and Guner, 2001a; Guner et al., 2004). As described previously, computational approaches for drug discovery and development may have more impact if integrated (Swaan and Ekins, 2005) and we have previously attempted to show that computational methods have been broadly applied to virtually all important proteins in absorption, distribution, metabolism, excretion and toxicity (ADME/Tox) (Ekins and Swaan, 2004b). The qaim of this paper is to provide an up-to-date review of all proteins and protein families addressed through current state-of-the-art in silico pharmacology methods.
Table 1.
A broad selection of in silico pharmacology targets that have been used with computational methods to discover new molecules with binding affinity
Abbreviations: AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazole propionate; COX, cyclooxygenase; CYP, cytochrome P450; HIV-1, human immunodeficiency virus; LOX, 5 lipoxygenase.
Drug target examples
Enzymes:The ubiquitin regulatory pathway, in which ubiquitin is conjugated and deconjugated with substrate proteins, represents a source of many potential targets for modulation of cancer and other diseases (Wong et al., 2003). The recent crystal structure of a mammalian de-ubiquitinating enzyme HAUSP, which specifically de-ubiquitinates the ubiquitinated p53 protein, may also assist in drug development despite the peptidic nature of its substrate (Hu et al., 2002). Novel non-peptidic inhibitors of the protease ubiquitin isopeptidase, which not only de-ubiquitinates p53 but other general ubiquitinated proteins as well, were discovered recently using a simple pharmacophore-based search of the National Cancer Institute (NCI) database (Mullally et al., 2001; Mullally and Fitzpatrick, 2002). These inhibitors had IC50 values in the low micromolar range and caused cell death independent of the tumour suppressor p53, which is mutated in greater than 50% of all cancers (hence, p53 inhibition per se may not represent an optimal target for modulation). The ubiquitin isopeptidase inhibitors shikoccin, dibenzylideneacetone, curcumin and the more recently described punaglandins from coral indicate that a sterically accessible α,β-unsaturated ketone is essential for bioactivity (Verbitski et al., 2004). All these molecules represent valuable leads for further chemical optimization.
Aromatase (cytochrome P450 (CYP)19) is a validated target for breast cancer. A ligand-based pharmacophore was generated with three non-steroidal inhibitors. This model could recognize known inhibitors from an in-house library and was further refined by the addition of molecular shape. The model was further used to search the NCI database and molecules were scored with a quantitative Catalyst Hypo-Refine (Accelrys Inc., San Diego, CA, USA) model generated with 16 molecules. The hits were also filtered with other pharmacophores for toxicity-related proteins, before testing. Two out of the three compounds were ultimately found to be micromolar inhibitors (Schuster et al., 2006).
A structure-based Catalyst pharmacophore was developed for acetylcholine esterase, which was subsequently used to search a natural product database. The strategy identified scopoletin and scopolin as hits and were later shown to have moderate in vivo activity (Rollinger et al., 2004). The same database was also screened against cyclooxygenase (COX)-1 and COX-2 structure-based pharmacophores, leading to the identification of known COX inhibitors. These represent examples where a combination of ethnopharmacological and computational approaches may aid drug discovery (Rollinger et al., 2005).
A combined ligand-based and structure-based approach was taken to gaining structural insights into the human 5-lipoxygenase (LOX). A Catalyst qualitative HipHop model was created with 16 different molecules that resulted in a five-feature pharmacophore. A homology model of the enzyme was based on two soybean LOX enzymes and one rabbit LOX enzyme. Molecular docking was then used to update and refine the pharmacophore to a four-feature model that could also be visualized in the homology model of 5-LOX. As a result of these models, amino-acid residues in the binding site were suggested as targets for site-directed mutagenesis while virtual screening with the pharmacophore had suggested compounds with a phenylthiourea or pyrimidine-5-carboxylate group for testing (Charlier et al., 2006). Homology models for the human 12-LOX and 15-LOX have also been used with the flexible ligand docking programme Glide (Schrödinger Inc.) to perform virtual screening of 50 000 compounds. Out of 20 compounds tested, 8 had inhibitory activity and several were in the low micromolar range (Kenyon et al., 2006).
More than 30 years of research on renin have not been enough to deliver a marketed drug that inhibits this enzyme. In spite of this, renin remains an attractive yet elusive target for hypertension (Fisher and Hollenberg, 2001; Stanton, 2003). In this respect, application of structure-based design leads to the identification of new non-peptidic inhibitors of human renin. These molecules include aliskiren (Rahuel et al., 2000; Torres et al., 2003), piperidines, including Ro-0661168 (Guller et al., 1999; Oefner et al., 1999; Vieira et al., 1999), and related 3,4-disubstituted piperidines (Marki et al., 2001). Interestingly, these piperidines bind to and stabilize a different conformer of the protein termed ‘open renin' (Bursavich and Rich, 2002), whereas aliskiren binds to ‘closed renin'. Since these latter structure-based design efforts, there have been remarkably very few published attempts at computer-aided design of novel renin inhibitors. A single early QSAR was derived for a series of chain-modified peptide analogues of angiotensinogen. The activity of these molecules was found to correlate with Kier's first-order molecular connectivity index descriptor and molecular weight but not with lipophilicity as measured by logP (Khadikar et al., 2005). Another computational method for renin drug discovery used the de novo design software GrowMol, which could apparently regenerate 3,4-disubstituted piperidines in 1% of the grown structures (Bursavich and Rich, 2002). An attempt to use a Catalyst pharmacophore to discover new renin inhibitors was described in the early 1990s (Van Drie, 1993). Several novel molecules from the Pomona database (an early three-dimensional (3D) molecule database) were found that mapped to a renin pharmacophore but apparently were not tested in vitro. More recently, a LigandFit docking study with a crystal structure of the ‘open renin' form was able to detect 10 known inhibitors seeded in a library of 1000 compounds within the top 8.4% when using a consensus scoring function. Four examples of high-scoring compounds that were not tested as inhibitors fulfilled the pharmacophore derived from the X-ray data, consisting of four hydrophobes, a hydrogen bond donor or positive ionizable feature as well as excluded volumes (Krovat and Langer, 2004). Another study has used similarity searching of the MDDR database (for over 100 000 compounds) using 10 renin inhibitors and was able to produce enrichment factors that were 17-fold greater than random (Hert et al., 2004). Genetic algorithms have also been used for class discrimination between renin inhibitors and non-inhibitors in a subset of the MDDR using a small number of interpretable descriptors. Among them, amide bond count, molecular weight and hydrogen bond donor counts were found to be much higher in renin inhibitors (Ganguly et al., 2006). The recent publications on novel renin inhibitors represent a considerable amount of new information that could be used for further QSAR model development and database searching efforts in order to derive novel starting scaffolds for optimization.
Cathepsin D is an aspartic protease found mainly in lysosomes, which may have a role in β-amyloid precursor protein release and hence may well be a target for Alzheimer's disease. Cathepsin D may also be elevated in breast cancer and ovarian cancer hence a means to modulate this activity could be beneficial in these diseases. There has been a brief overview of Cathepsin D in a comprehensive review of protease inhibitors (Leung et al., 2000). A combination of a structure-based design algorithm and combinatorial chemistry has been successfully applied to finding novel molecules for Cathepsin D in the nanomolar range (Kick et al., 1997). Structures based on pepstatin (a 3.8 pM inhibitor (Baldwin et al., 1993)) yielded a 6–7% hit rate. These molecules were tested in vitro using hippocampal slices and were shown to block the formation of hyperphosphorylated Tau fragments (Bi et al., 2000). There have been relatively few computational studies to date on Cathepsin D and other related aspartic proteases such as renin and β-secretase. One study has used molecular dynamics and free energy analyses (MM-PBSA) of Cathepsin D inhibitor interactions to suggest new substitutions that may improve binding (Huo et al., 2002). A genetic algorithm-based de novo design tool, ADAPT has also been used to rediscover active Cathepsin D molecules, by placing key fragments in the correct positions (Pegg et al., 2001). Computational models may aid in the selection of novel ligands for protease inhibition that are non-peptidic and selective. Using the structural features of eight published inhibitors for Cathepsin D (Huo et al., 2002), a five-feature pharmacophore was derived consisting of three hydrophobes and two hydrogen bond acceptors (r=0.98). This pharmacophore was used to search a molecule database and selected 10 molecules out of 11 441 present. In contrast, a similarity search at the 95% level using ChemFinder (CambridgeSoft, Cambridge, MA, USA) suggested 16 different molecules. All of these were selected for testing in vitro. The pharmacophore produced four hits (40% hit rate) and the similarity search generated five hits (31% hit rate), where at least one replicate showed greater than 40% inhibition (Ekins et al., 2004a). In silico evaluation of the ADME properties for all active compounds estimated that the molecules would be well absorbed, although some were predicted to have solubility and CYP2D6 inhibition problems.
Pharmacophore- and structure-based approaches have been used to optimize an acyl urea hit for human glycogen phosphorylase. A Catalyst HypoGen five-feature pharmacophore was developed and used to guide further analogue synthesis. These compounds showed a good correlation with prediction (r=0.71). An X-ray structure for one molecule was used to confirm the predicted binding conformation. Ultimately, a comparative molecular field analysis (CoMFA) model was generated with all molecules synthesized and was found to be complementary to the X-ray structure. The outcome of this study was a molecule with good cellular activity that could inhibit blood glucose levels in vivo in rat (Klabunde et al., 2005).
The human sirtuin type 2, a target for controlling aging and some cancers, deacetylates α-tubulin and has been crystallized at high resolution. This structure has been used for docking the Maybridge database and returned a small hit list from which 15 compounds were tested and 5 showed activity at the micromolar level (Tervo et al., 2004).
Catechol O-methyltransferase is a target for Parkinson's disease and there is currently a crystal structure of the enzyme that has been used to generate a homology model of the human enzyme. This model was used to dock with FlexX software several catechins from tea and understand the structure-activity relationship (SAR) for these molecules and their metabolites, which had been tested in vitro. Ultimately, the combination of in vitro and computational work indicated that the galloyl group on catechins, the distance between Lys 144 on the enzyme, and the reacting catecholic hydroxy group were important for inhibition (Chen et al., 2005).
Kinases:
The kinases represent an attractive family of over 500 targets for the pharmaceutical industry, with several drugs approved recently. Kinase space has been mapped using selectivity data for small molecules to create a chemogenomic dendrogram for 43 kinases that showed the highly homologous kinases to be inhibited similarly by small molecules (Vieth et al., 2004). Virtual screening methods have been applied quite widely for kinases to date (Fischer, 2004). The structure-based design method has produced new potent inhibitors of CDK1 starting from the highly similar apo CDK2 and the positioning of olomoucine. A few amino-acid residues were mutated to conform to the CDK1 sequence. MacroModel was used to energy minimize molecules in the ATP pocket and visual inspection suggested points for molecular modification on the ligand. Very quickly, design efforts guided ligand optimization to improve activity from 4.5 μM to 25 nM (Furet et al., 2000). A more recent CDK1/cyclin B homology model was also used to manually dock ligands, which enabled progression from alsterpaullone with an IC50 of 35 nM to a derivative with an IC50 of 0.23 nM (Kunick et al., 2005).
A structure-based in silico screening method was pursued for the Syk C-terminal SH2 domain using DOCK to find low molecular weight fragments for each binding site with millimolar binding affinity. The fragments were then linked to result in molecules in the 38–350 μM range, which is a starting point for further lead optimization (Niimi et al., 2001).
A pseudoreceptor model was built with a set of 27 epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors with the flexible atom receptor model method. The top 15 models created had high r2 and q2 and were also validated with a six-molecule test set. The pseudoreceptor was also in accord with a crystal structure of CDK2 (Peng et al., 2003).
Virtual screening using DOCK with the crystal structure of the Lck SH2 domain was used to screen two million commercially available molecules. Extensive filtering was required to result in a manageable hit list using molecular weight and diversity. Out of 196 compounds tested in vitro, 34 were inhibitory at 100 μM, while 2 had activities of 10 and 40 μM. Fluorescence titrations of some of these compounds suggested the KD values were in the low micromolar range (Huang et al., 2004). The same group also took a similar approach to discover inhibitors of ERK2 by screening 800 000 compounds computationally and testing in vitro 80 of them (Hancock et al., 2005). Five of these molecules inhibited cell proliferation and two were shown by fluorescence titration to bind ERK2 with KD values, which were in the low micromolar range. In both cases, docking of the active molecules suggested orientations for verification by X-ray crystallography (Hancock et al., 2005).
The Ligand Scout method was used with BCR-ABL tyrosine kinase to find STI-571 (imatinib, Gleevec) in a single and multiple conformation database (Wolber and Langer, 2005). A structurally related three-substituted benzamidine derivative of STI-571 was suggested by structure-based design and when manually docked into the binding site and energy minimized, it was shown to form favourable interactions with a hydrophobic pocket.
CK2 and PKD are part of the COP9 signalosome and can control stability of p53 and c-Jun, which are important for tumour development. Curcumin, besides being an inhibitor of ubiquitin isopeptidase (Mullally et al., 2001; Mullally and Fitzpatrick, 2002) and activator protein-1 (Tsuchida et al., 2006), also inhibits CK2 and PKD. Using curcumin and emodin as reference structures against which a database of over a million molecules was screened by means of 2D and 3D similarity searches retrieved 35 molecules. Among them, seven possessed inhibitory activity. For example, piceatannol was more potent than curcumin against both CK2 and PKD, with IC50 values of 2.5 and 0.5 μM, respectively (Fullbeck et al., 2005). Obviously, these examples suggest there has been some success in finding active molecules for kinases, but interestingly in few of these studies is selectivity toward other kinases accounted for. Ultimately, for therapeutic success activity toward several kinases (but selectivity toward others) may be required.
Drug-metabolizing enzymes and transporters:
Mathematical models describing quantitative structure–metabolism relationships were pioneered by Hansch et al. (1968) using small sets of similar molecules and a few molecular descriptors. Later, Lewis and co-workers provided many QSAR and homology models for the individual human CYPs (Lewis, 2000). As more sophisticated computational modelling tools became available, we have seen a growth in the number of available models (de Groot and Ekins, 2002b; de Graaf et al., 2005; de Groot, 2006) and the size of the data sets they encompass. Some more recent methods are also incorporating water molecules into the binding sites when docking molecules into these enzymes and these may be important as hydrogen bond mediators with the binding site amino acids (Lill et al., 2006). Docking methods can also be useful for suggesting novel metabolites for drugs. A recent example used a homology model of CYP2D6 and docked metoclopramide as well as 19 other drugs to show a good correlation between IC50 and docking score r2=0.61 (Yu et al., 2006). A novel aromatic N-hydroxy metabolite was suggested as the major metabolite and confirmed in vitro. Now that several crystal structures of the mammalian CYPs are available, they have been found to compare quite favourably to the prior computational models (Rowland et al., 2006). However, for some enzymes like CYP3A4, where there is both ligand and protein promiscuity, there may be difficulty in making reliable predictions with some computational approaches such as docking with the available crystal structures (Ekroos and Sjogren, 2006). Hence, multiple pharmacophores or models may be necessary for this and other enzymes (Ekins et al., 1999a, 1999b), as it has been indicated by others more recently (Mao et al., 2006).
The UDP-glucuronosyltransferases are a class of versatile enzymes involved in the elimination of drugs by catalysing the conjugation of glucuronic acid to substrates bearing a suitable functional group, so called phase II enzymes. There have been numerous QSAR and pharmacophore models that have been generated with relatively small data sets for rat and human enzymes. The pharmacophores for the human UGT1A1, UGT1A4 and UGT1A9 all have in common two hydrophobes and a glucuronidation feature, while UGT1A9 has an additional hydrogen bond acceptor feature (Smith et al., 2004; Sorich et al., 2004). Sulfotransferases, a second class of conjugating enzymes, have been crystallized (Dajani et al., 1999; Gamage et al., 2003) and a QSAR method has also been used to predict substrate affinity to SULT1A3 (Dajani et al., 1999). To the best of our knowledge, computational models for other isozymes have not been developed. In general, conjugating enzymes have generally been infrequently targeted for in silico models. Perhaps because of a paucity of in vitro data and limited diversity of molecules tested, they have been less widely applied in industry.
The computational modelling of drug transporters has been thoroughly reviewed by numerous groups (Zhang et al., 2002a, 2002b; Chang and Swaan, 2005) and will not be addressed here in detail. Various transporter models have also been applied to database searching to discover substrates and inhibitors (Langer et al., 2004; Pleban et al., 2005; Chang et al., 2006b) and increase the efficiency of in vitro screening (Chang et al., 2006a) or enrichment over random screening. A pharmacophore model of the Na+/D-glucose co-transporter found in renal proximal tubules was derived indirectly using phlorizin analogues with the DISCO programme to superpose molecules. This enabled an estimate of the size of the binding site to be obtained. In contrast to more recent studies with transporter pharmacophores, this model was not tested or used for database searching (Wielert-Badt et al., 2000).
Receptors:
There are more than 20 different families of receptors that are present in the plasma membrane, altogether representing over 1000 proteins of the receptorome (Strachan et al., 2006). Receptors have been widely used as drug targets and they have a wide array of potential ligands. However, it should be noted that to date we have only characterized and found agonists and antagonists for a small percentage of the receptorome. The α-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor is central to many central nervous system (CNS) pathologies and ligands have been synthesized as anticonvulsants and neuroprotectants. There is currently no 3D structure information and therefore a four-point Catalyst HIPHOP pharmacophore was developed with 14 antagonists. This was then used to search the Maybridge database and select eight compounds for testing of which six of these were found to be active in vivo as anticonvulsants (Barreca et al., 2003).
Serotonin plays a role in many physiological systems, from the CNS to the intestinal wall. Along with its many receptors, it has a major developmental function regulating cardiovascular morphogenesis. The 5-HT2 receptor family are G protein-coupled 7-transmembrane spanning receptors with 5-HT2B expressed in cardiovascular, gut, brain tissues, as well as human carcinoid tumors (Nebigil et al., 2000). In recent years, this receptor has been implicated in the valvular heart disease defects caused by the now banned ‘fen-phen' treatment of patients. The primary metabolite, norfenfluramine, potently stimulates 5-HT2B (Fitzgerald et al., 2000; Rothman et al., 2000). Computational modelling of this receptor has been limited to date. A traditional QSAR study used a small number of tetrahydro-β-carboline derivatives as antagonists of the rat 5-HT2B contractile receptor in the rat stomach fundus (Singh and Kumar, 2001). A 3D-QSAR with GRID-GOLPE using 38 (aminoalkyl)benzo and heterocycloalkanones as antagonists of the human receptor resulted in very poor model statistics, possibly owing to the limited range of activity measured and the fact that the data corresponded to a functional response that is likely more complex (Brea et al., 2002). Neither of these models was validated with external predictions. On the basis of bacteriorhodopsin and rhodopsin, homology models for the mouse and human 5-HT2B receptor have been combined with site-directed mutagenesis. The bacteriorhodopsin structure provided more reliable models, which confirmed an aromatic box hypothesis for ligand interaction along transmembrane domains 3, 6, 7 with serotonin (Manivet et al., 2002). A more recent 5-HT2B homology model based on the rhodopsin-based model of the rat 5-HT2A was used to determine the sites of interaction for norfenfluramine following molecular dynamics simulations. Site-directed mutagenesis showed that Val 2.53 was implicated in high-affinity binding through van der Waals interactions and the ligand methyl groups (Setola et al., 2005). There is certainly an opportunity to develop further QSAR models for this receptor in order to rapidly screen libraries of molecules to identify undesirable potent inhibitors.
The serotonin 5-HT1A receptor has been frequently modelled. For example, a conformational study of four ligands defined a pharmacophore of the antagonist site using SYBYL (Hibert et al., 1988). The model resulting from such an active analogue approach was used in molecule design and predicted molecule stereospecificity. More recently, a series of over 700 homology models were iteratively created based on the crystal structure of the bovine rhodopsin that were in turn tuned by FlexX docking of known ligands. The final model was used in a virtual screening simulation that was enriched with inhibitors, compared with random selection and from this the authors suggested its utility for a real virtual screen (Nowak et al., 2006). A homology model of the 5-HT1A receptor has also been used with DOCK to screen a library of 10 000 compounds seeded with 34 5-HT1A ligands. Ninety percent of these active compounds were ranked in the top 1000 compounds (Becker et al., 2006), representing a significant enrichment. The same model was used to screen a library of 40 000 vendor compounds and select 78 for testing, of which 16 had activities below 5 μM, one possessing 1 nM affinity. Structure-based in silico optimization was then performed to improve selectivity with other GPCRs and optimize the pharmacokinetic (PK) profile. However, as this proceeded, the molecules were found to have affinity for the human ether a-go-go-related gene (hERG), and this was subsequently computationally assessed using a homology model that pointed to adjusting the hydrophobicity. The resulting clinical candidate had good target and antitarget selectivity and backup compounds were selected in the same way (Becker et al., 2006).
Another early computer-aided pharmacophore generated with SYBYL using a set of selective and non-selective analogues was used to design agonists for 5-HT1D as antimigraine agents with selectivity against 5-HT2A (linked to undesirable changes in blood pressure) (Glen et al., 1995).
A range of typical and atypical antipsychotics bind to the 5-HT6 receptor. Based on the structure of bovine rhodopsin, homology models of the human and rodent 5-HT6 receptors were constructed and used to dock ligands that were known to exhibit species differences in binding (Hirst et al., 2003). Following sequence alignment, amino-acid residues were identified for mutation and the rationalization of these mutations and their effects on ligand binding were obtained from the docking studies. The models generated were in good agreement with the in vitro data and could be used for further molecule design. This study was a good example where computational, molecular biology and traditional pharmacology methods were combined (Hirst et al., 2003).
The Na+, K+-ATPase is a receptor for cardiotonic steroids, which in turn inhibit the ATPase and cation transport and have ionotropic actions. Although the effects of digitalis have been known for hundreds of years, a molecular understanding has remained absent until recently. A homology model was generated with the SERCA1a crystal structure and tested with nine cardiac glycosides (Keenan et al., 2005). The model was also mutated to mimic the rat receptor and showed how oubain would orient differently in these models, perhaps explaining the species difference in affinity. These models also suggested amino acids that could be experimentally mutated to validate the hypothesis for the binding site identification, although this has yet to be tested.
The dopamine receptors have been implicated in Parkinson's disease and schizophrenia. Unfortunately, no crystal structure is currently available and thus the search for new antagonists has used QSAR models. A set of 48 compounds was used with four different QSAR methods (CoMFA, simulated annealing-partial least square (PLS), k-nearest neighbours (kNN) and SVM), and training as well as testing statistics were generated. SVM and kNN models were also used to mine compound databases of over 750 000 molecules that resulted in 54 consensus hits. Five of these hits were known to bind the receptor and were not in the training set, while other suggested hits did not contain the catechol group normally seen in most dopamine inhibitors (Oloff et al., 2005).
The α1A receptor is a target for controlling vascular tone and therefore useful for antihypertensive agents. A novel approach for ligand-based screening called multiple feature tree (MTree) describes the training set molecules as a feature tree descriptor derived from a topological molecular graph that is then aligned in a pairwise fashion (Hessler et al., 2005). A set of six antagonists was used to derive a model with this method and was compared with a Catalyst pharmacophore model. Both approaches identified a central positive ionizable feature flanked by hydrophobic regions at either end. These two methods were compared for their ability to rank a database of over 47 000 molecules. Within the top 1% of the database, MTree had an enrichment factor that was over twice that obtained with Catalyst (Hessler et al., 2005).
Nuclear receptors:
Nuclear receptors constitute a family of ligand-activated transcription factors of paramount importance for the pharmaceutical industry since many of its members are often considered as double-edged swords (Shi, 2006). On the one hand, because of their important regulatory role in a variety of biological processes, mutations in nuclear receptors are associated with many common human diseases such as cancer, diabetes and osteoporosis and thus, they are also considered highly relevant therapeutic targets. On the other hand, nuclear receptors act also as regulators of some the CYP enzymes responsible for the metabolism of pharmaceutically relevant molecules, as well as transporters that can mediate drug efflux, and thus they are also regarded as potential therapeutic antitargets (off-targets).
Examples of the use of target-based virtual screening to identify novel small molecule modulators of nuclear receptors have been recently reported. Using the available structure of the oestrogen receptor subtype α (ERα) in its antagonist conformation, a homology model of the retinoic acid receptor α (RARα) was constructed (Schapira et al., 2000). Using this homology model, virtual screening of a compound library lead to the identification of two novel RARα antagonists in the micromolar range. The same approach was later applied to discover 14 novel and diverse micromolar antagonists of the thyroid hormone receptor (Schapira et al., 2000). By means of a procedure designed particularly to select compounds fitting onto the LxxLL peptide-binding surface of the oestrogen receptor, novel ERα antagonists were identified (Shao et al., 2004). Since poor displacement of 17β-estradiol was observed in the ER-ligand competition assay, these compounds may represent new classes of ERα antagonists, with the potential to provide an alternative to current anti-oestrogen therapies. The discovery of three low micromolar hits for ERβ displaying over 100-fold binding selectivity with respect to ERα was also recently reported using database screening (Zhao and Brinton, 2005). A final example reports the identification and optimization of a novel family of peroxisome proliferator-activated receptors-γ partial agonists based upon pyrazol-4-ylbenzenesulfonamide after employing structure-based virtual screening, with good selectivity profile against the other subtypes of the same nuclear receptor group (Lu et al., 2006).
Ion channels:
Therapeutically important channels include voltage-gated ion channels for potassium, sodium and calcium that are present in the outer membrane of many different cells such as those responsible for the electrical excitability and signalling in nerve and muscle cells (Terlau and Stuhmer, 1998). These represent validated therapeutic targets for anaesthesia, CNS and cardiovascular diseases (Kang et al., 2001). A recent review has discussed the various QSAR methods such as pharmacophores, CoMFA, SVM, 2D-QSAR, Genetic Programming, Self Organizing Maps and recursive partitioning that have been applied to most ion channels (Aronov et al., 2006) in the absence of crystal structures. To date L-type calcium channels and hERG appear to have been the most extensively studied channels in this regard. In contrast, there are far fewer examples of computational models for the sodium channel. These three classes of ion channels have been studied as they represent either therapeutic targets or antitargets to be avoided.
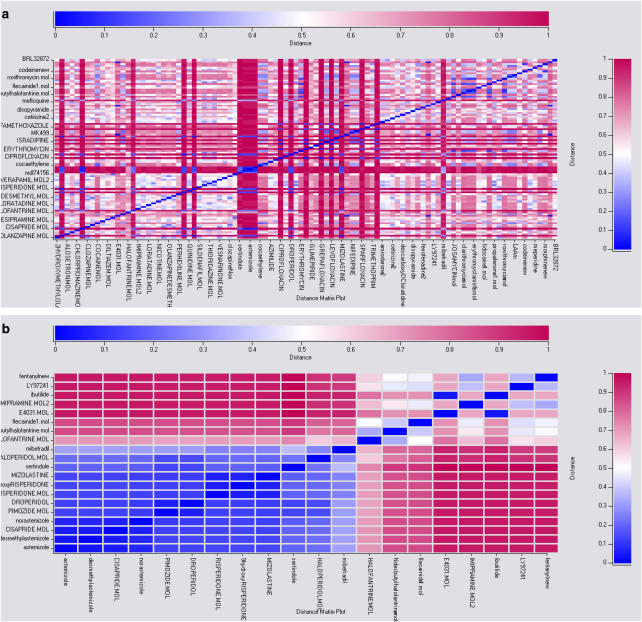
For example, one of many models for the hERG potassium channel has compared three different methods with the same set of molecules for training and a test set. Recursive partitioning, Sammon maps and Kohonen maps were used with atom path lengths (Ekins et al., 2006). The average classification quality was high for both training and test selections. The Sammon mapping technique outperformed the Kohonen maps in classification of compounds from the external test set. The quantitative predictions for recursive partitioning could be filtered using a Tanimoto similarity to remove molecules that were markedly different to the training set (Willett, 2003). The path length descriptors can also be used to visualize the similarity of the molecules in the whole training set (Figure 1a). In addition, a subset of molecules can also be compared, with those highlighted in blue representing close neighbours and those in red being more distant (Figure 1b).
Figure 1.
(a) A distance matrix plot of the 99 molecule hERG training set showing in general that the molecules are globally dissimilar as the plot is primarily red (Ekins et al., 2006). (b) A distance matrix plot of a subset of the training set to show molecules similar to astemizole. Blue represents close molecules and red represents distant molecules based on the ChemTree pathlength descriptors (see colour scale).
Transcription factors:
A cyclic decapeptide with activity against the AP-1 transcription factor was used to derive a 3D pharmacophore to which low energy conformations of non-peptidic compounds were compared. New 1-thia-4-azaspiro[4,5]decane and benzophenone derivatives with activity in binding and cell-based assays were discovered as AP-1 inhibitors in a lead hopping approach (Tsuchida et al., 2006).
Antibacterials:
Twenty deoxythymidine monophosphate analogues were used along with docking to generate a pharmacophore for Mycobacterium tuberculosis thymidine monophosphosphate kinase inhibitors with the Catalyst software. A final model was used to screen a large database spiked with known inhibitors. The model was suggested to have an enrichment factor of 17, which is highly significant. In addition, the model was used to rapidly screen half a million compounds in an effort to discover new inhibitors (Gopalakrishnan et al., 2005).
Antivirals:
Neuroamidase is a major surface protein in influenza virus. A structure-based approach was used to generate Catalyst pharmacophores and these in turn were used for a database search and aided the discovery of known inhibitors. The hit lists were also very selective (Steindl and Langer, 2004).
Human rhinovirus 3C protease is an antirhinitis target. A structure-based pharmacophore was developed initially around AG 7088 but this proved too restrictive. A second pharmacophore was developed from seven peptidic inhibitors using the Catalyst HIPHOP method. This hypothesis was useful in searching the world drug index database to retrieve compounds with known antiviral activity and several novel compounds were selected from other databases with good fits to the pharmacophore, indicative that they would be worth testing although these ultimate testing validation data were not presented (Steindl et al., 2005a). Human rhinovirus coat protein is another target for antirhinitis. A combined pharmacophore, docking approach and PCA-based clustering was used. A pharmacophore was generated from the structure and shape of a known inhibitor and tested for its ability to find known inhibitors in a database. Ultimately, after screening the Maybridge database, 10 compounds were suggested that were then docked and scored. Six compounds were tested and found to inhibit viral growth. However, the majority of them were found to be cytotoxic or had poor solubility (Steindl et al., 2005b). The Ligand Scout approach was tested on the rhinovirus serotype 16 and was able to find known inhibitors in the PDB (Wolber and Langer, 2005).
The SARS coronavirus 3C-like proteinase has been addressed as a potential drug design target. A homology model was built and chemical databases were docked into it. A pharmacophore model and drug-like rules were used to narrow the hit list. Forty compounds were tested and three were found with micromolar activity, the best being calmidazolium at 61 μM (Liu et al., 2005), perhaps a starting point for further optimization.
A pharmacophore has also been developed to predict the hepatitis C virus RNA-dependent RNA polymerase inhibition of diketo acid derivatives. A Catalyst HypoGen model was derived with 40 molecules with activities over three log orders to result in a five-feature pharmacophore model. This was in turn tested with 19 compounds from the same data set as well as nine diketo acid derivatives, for which the predicted and experimental data were in good agreement (Di Santo et al., 2005).
Other therapeutic targets:
The integrin VLA-4 (α4β1) is a target for autoimmune and inflammatory diseases such as asthma and rheumatoid arthritis. The search for antagonists has included using a Catalyst pharmacophore derived from the X-ray crystal structure of a peptidic inhibitor (Singh et al., 2002b). This was used to search a virtual database of compounds that could be made with reagents from the available chemicals directory. Twelve compounds were then selected and synthesized, with resulting activities in the range between 1.3 nM and 20 μM. Hence, a peptide was used to derive non-peptide inhibitors that were active in vivo. A second study by the same group used CoMFA with a set of 29 antagonists with activity from 1 to 662 nM to generate a model with good internal validation statistics that was subsequently used to indicate favourable regions for molecule substituent changes (Singh et al., 2002a). It is unclear whether the CoMFA model was also successful for design of further molecules.
It is possible to use approved drugs as a starting point for drug discovery for other diseases. For example, the list of World Health Organization essential drugs has been searched to try to find leads for prion diseases using 2D Tanimoto similarity or 3D searching with known inhibitors. This work to date has suggested compounds, yet they appear not to have been tested, so the approach has not been completely validated (Lorenzen et al., 2005).
Protein–protein interactions are key components of cellular signalling cascades, the selective interruption of which would represent a sought after therapeutic mechanism to modulate various diseases (Tesmer, 2006). However, such pharmacological targets have been difficult for in silico methods to derive small molecule inhibitors owing to generally quite shallow binding sites. The G-protein Gβγ complex can regulate a number of signalling proteins via protein–protein interactions. The search for small molecules to interfere with the Gβγ-protein–protein interaction has been targeted using FlexX docking and consensus scoring of 1990 molecules from the NCI diversity set database (Bonacci et al., 2006). After testing 85 compounds as inhibitors of the Gβ1γ2-SIRK peptide, nine compounds were identified with IC50 values from 100 nM to 60 μM. Further substructure searching was used to identify similar compounds to one of the most potent inhibitors to build a SAR. These efforts may eventually lead to more potent lead compounds.
Complex property modelling
Up to this point, we have generally considered in silico pharmacology models that essentially relate to a single target protein and either the discovery of molecules as agonists, antagonists or with other biological activity after database searching and in vitro testing or following searching of databases seeded with molecules of known activity for the target. However, there are many complex properties that have been modelled in silico and these will be briefly discussed here. It should also be pointed out that while several physicochemical properties such as ClogP and water solubility have been extensively studied, the training sets for these models are in the 1000s or tens of thousands of molecules, while other complex properties have generally used much smaller training sets in the range of hundreds of molecules.
For example, a measure of molecule clearance would be indicative of elimination half-life that would naturally be of value for selecting candidates. The intrinsic clearance has therefore been used as a measure of the enzyme activity toward a compound and this may involve multiple enzymes. Some of the earliest models for this property includes a CoMFA model of the CYP-mediated metabolism of chlorinated volatile organic compounds, likely representative of CYP2E1 (Waller et al., 1996). A more generic set of molecules with clearance data derived from human hepatocytes has been used to predict human in vivo clearance using multiple linear regression, PCA, PLS, Neural Networks with leave-one-out cross-validation (Schneider et al., 1999). Microsomal and hepatocyte clearance data sets have also been used separately to generate Catalyst pharmacophores, which were then tested by predicting the opposing data set. This method assumes there are some pharmacophore features intrinsic to the molecules that dictate intrinsic clearance (Ekins and Obach, 2000).
A second complex property is the volume of distribution that is a function of the extent of drug partitioning into tissue versus plasma and there have been several attempts at modelling this property (Lombardo et al., 2002, 2004). This property, along with the plasma half-life, determines the appropriate dose of a drug. For example, 253 diverse drugs from the literature were used with eight molecular descriptors with Sammon and Kohonen mapping methods. These models appeared to classify correctly 80% of the compounds (Balakin et al., 2005). Recently, a set of 384 drugs with literature volume of distribution at steady-state data was used with a mixture discriminant analysis-random forest method and 31 molecular descriptors to generate a predictive model. This model was tested with 23 molecules, resulting in a geometric mean fold error of 1.78, which was comparable to the values for other predictions for this property from animal, in vitro, or other methods (Lombardo et al., 2006).
A third property, the plasma half-life determined by numerous ADME properties has also been modelled with Sammon and Kohonen maps using data for 458 drugs from the literature and four molecular descriptors. Like the previously described volume of distribution models, these models appeared to classify correctly 80% of the compounds (Balakin et al., 2005).
A fourth complex property is renal clearance, which assumes the excretion of the unchanged drug that takes place only by this route, hence this represents a method of monitoring the proportion of drug metabolized. In one set of published QSAR models, 130 molecules were used with 62 Volsurf or 37 Molconn-Z descriptors. The models were tested with 20 molecules and one using soft independent modelling of class analogies and Molconn-Z descriptors obtained 85% correct classification between the two classes (0–20 and 20–100%) (Doddareddy et al., 2006).
A fifth example of a complex property is the protein–ligand interaction and appropriate scoring functions for which several methods have been developed such as force fields, empirical and knowledge-based approaches (see also Ekins et al., 2007). These are important in computational structure-based design methods for assessing virtual candidate molecules to select those that are likely to bind a protein with highest affinity (Shimada, 2006). Recently, a Kernel partial least squares (K-PLSs) QSAR approach has been used along with a genetic algorithm feature selection method for the distance-dependent atom pair descriptors from the 61 or 105 small molecule training sets with binding affinity data and the proteins they bind to. Bootstrapping, scrambling the data and external test sets were used to test the models (Deng et al., 2004). In essence, such K-PLS QSAR models across many proteins perhaps isolate the key molecular descriptors that relate to the highest affinity interactions. It will be interesting to see whether such models can continue to be generated with the much larger binding affinity data sets that are now available.
A final example of a complex property is the Vmax of an enzyme that has been modelled on a few occasions (Hirashima et al., 1997; Mager et al., 1982; Ghafourian and Rashidi, 2001; Sipila and Taskinen, 2004). This value will depend on the properties of the compound in question and will be influenced by the steric properties of the active site as well as the ease of expulsion of the leaving group from the active site. Balakin et al. (2004), have recently used neural network methods to model the Vmax data for N-dealkylation mediated by CYP2D6 and CYP3A4, using whole molecules, centroid of the reaction and leaving group-related descriptors. These models were also used to predict small sets of molecules not included in training. Ultimately, many other reactions and the evaluation of other enzymes will be necessary. Similarly, larger test sets are required for all the above complex property models to provide further confidence in the models in terms of their utility and applicability.
Current scope, limitations, and trends
Uses of in silico pharmacology
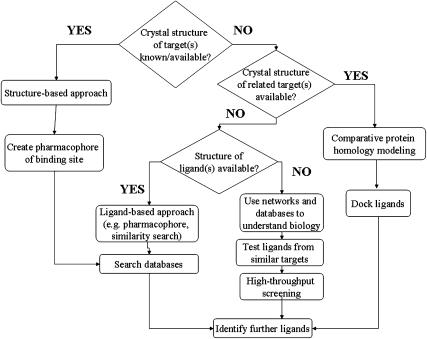
We propose a general schema for in silico pharmacology, which is shown in Figure 2. This demonstrates some of the key roles of the computational technologies that can assist pharmacology. These roles include finding new antagonists or agonists for a target using an array of methods either in the absence or presence of a structure for the target. Computational methods may also aid in understanding the underlying biology using network/pathways based on annotated data (signalling cascades), determining the connectivity of drug as a network with targets to understand selectivity, integration with other models for PK/PD (pharmacodynamic) and ultimately the emergence of systems in silico pharmacology. Obviously, we have taken more of a pharmaceutical bias in this review but we would argue these methods are equally amenable and should be considered to discover new chemical probes for the academic pharmacologist as opposed to lead molecules for optimization to become drugs. Some of the advantages of in silico pharmacology and in silico methods in general are the reduction in the number of molecules made and tested through database searching to find inhibitors or substrates, increased speed of experiments through reliable prediction of most pharmaceutical properties from molecule structure alone and ultimately reductions in animal and reagent use. We must however consider the multiple optimization of numerous predicted properties, possibly either weighting in silico pharmacology models by importance (or confidence in the model and or data), as well as data set size and diversity. Similarly, we should consider the disadvantages of in silico pharmacology methods as protein flexibility, molecule conformation and promiscuity all hinder accurate predictions. For example, even with the recent availability of crystal structures for several mammalian drug-metabolizing enzymes, there is still considerable difficulty in reliable metabolism predictions. Our focus thus far has been on the creation of many in silico pharmacology models for human properties, yet as pharmacology uses animals for much in vivo testing and subcellular preparations from several species for in vitro experiments, we need models from other species both to understand differences as well as enable better scaling between them. A widely discussed disadvantage of in silico methods is the applicability of the model, which will now be discussed further.
Figure 2.
A schematic for in silico pharmacology.
Defining in silico model applicability domain
Some of the in silico pharmacology methods that can be used have similar limitations to models used in other areas, such as those for predicting physicochemical and ADME/Tox properties. For example, models may be generated with a narrow homologous series of pharmacologically relevant molecules (local model) or a structurally diverse range of molecules (global model). These two approaches have their pros and cons, respectively. The applicability domain of the local model may be much narrower than for the global model such that changing to a new chemical series will result in prediction failure. However, global models may also fail if the predicted molecule falls far enough away from representative molecules in the training set. These limitations are particularly specific to QSAR models. From many of the in silico pharmacology model examples described above, the QSAR models are generally local in nature and this will limit lead hopping to new structural series, whereas global models may be more useful for this feature. Several papers have described the applicability domain of models and methods in considerable detail (Dimitrov et al., 2005; Tetko et al., 2006) to calculate this property. Molecular similarity to training set compounds may be a reliable measure for prediction quality (Sheridan et al., 2004) as demonstrated for a hERG model (Ekins et al., 2006). To our knowledge, there has not been a specific analysis of the applicability domain specifically for in silico pharmacology models (other than for those examples described above) to the same degree as there has been for physicochemical properties like solubility and logP. The applicability domain of pharmacophore models have not been addressed either as the focus has primarily been on statistical QSAR methods.
As we shift toward hybrid or meta-computational methods (that integrate several modelling approaches and algorithms) for predicting from molecular structure the possible physicochemical and pharmacological properties, then these could be used to provide prediction confidence by consensus. The docking methods with homology models for certain proteins of pharmacological interest could be used alongside QSAR or pharmacophore models if these are also available. There have been numerous occasions in the study of drug-metabolizing enzymes were QSAR and homology models have been combined or used to validate each other (de Groot et al., 2002a; de Graaf et al., 2005; de Groot, 2006).
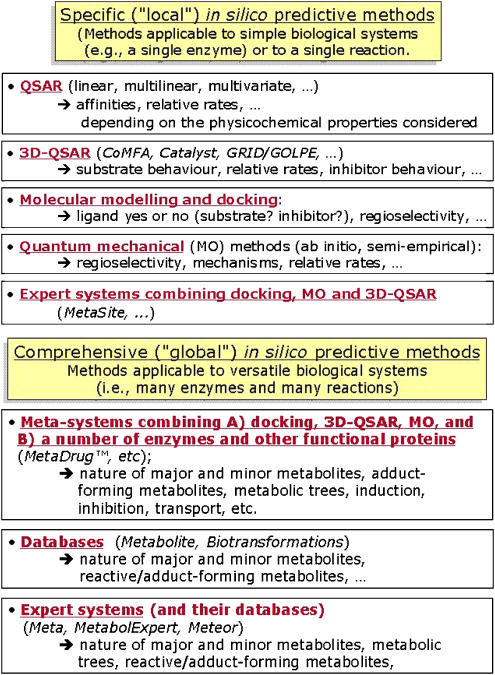
Drug metabolism is a good example as several simultaneous outcomes (for example, metabolites) often occur, a condition not normally found in other pharmacological assays where a single set of conditions yields a single outcome. It is here that the classification into specific (‘local') and comprehensive (‘global') methods finds its clearest use (see Figure 3), with local methods being applicable to simple biological systems such as a single enzyme or a single enzymatic activity (Testa and Krämer, 2006). The production of regioselective metabolites (for example, hydroxylation to a phenol and an alcohol) is usually predictable from such methods, but that of different routes (for example, oxidation versus glucuronidation) is not. This is where global algorithms (that is, applicable to versatile biological systems) are most useful in their potential capacity to encompass all or most metabolic reactions and offer predictions, which are much closer to the in vivo situation.
Figure 3.
Local and global models applied to drug metabolism. Figures are taken from Testa and Krämer (2006) with permission.
Observations and caveats
It is readily apparent that in a minority of papers we have found that computational approaches have resulted in predicted lead compounds for testing without the authors providing further experimental verification of biological activity (Krovat and Langer, 2004; Langer et al., 2004; Steindl and Langer, 2004; Gopalakrishnan et al., 2005; Lorenzen et al., 2005; Steindl et al., 2005a; Amin and Welsh, 2006). This is an interesting observation as for many years computational studies were generally performed after synthesis of molecules, and essentially provided illustrative pictures and explanation of the data. Now it appears we are seeing a shift in the other direction as predictions are published for pharmacological activity without apparently requiring in vitro or in vivo experimental verification, as long as the models themselves are validated in some manner. As the models may only have a limited prediction domain so perhaps in future we will see some discussion of the predicted molecules and their distance from the training set or some other measure of how far the predictions can be extended.
Many of the molecules identified by virtual screening techniques have not been tested in vitro to ensure that they are not false positives that may actually be involved in molecule aggregation. These types of molecules have been termed so-called ‘promiscuous inhibitors', occurring as micromolar inhibitors of several proteins (McGovern et al., 2002; McGovern and Shoichet, 2003; Seidler et al., 2003). A preliminary computational model was developed to help identify these potential promiscuous inhibitors (Seidler et al., 2003). From reviewing the literature, we suggest it would be worth researchers either implementing filters for ‘promiscuous inhibitors' or performing rigorous experimental verification of their predicted bioactive molecules to rule out this possibility.
Publication bias perhaps also limits the number of examples of failures of computational methods that are published (if any). It would certainly be very useful to know the existence of difficult targets for modelling with different methods, as this apparently is a process of trial and error for each investigator currently.
In summary, in this and the accompanying review (Ekins et al., 2007), we have presented our interpretation of in silico pharmacology and described how the field has developed so far and is used for: discovery of molecules that bind to many different targets and display bioactivity, prediction of complex properties and the understanding of the underlying metabolic and network interactions. While we have not explicitly discussed PK/PD, whole organ, cell or disease simulations in this review, we recognize they too are an important component of the computer-aided drug design approach (Noble and Colatsky, 2000; Gomeni et al., 2001; Kansal, 2004) and may be more widely integrated with other in silico pharmacology methods described previously (Ekins et al., 2005).
Conclusion
The brief history of in silico pharmacology has taken perhaps a rather predictable route with computational models applied to many of the most important biological targets where they have the capacity to be used to search large databases and quickly suggest molecules for testing. Many of the examples we have presented have demonstrated significant enrichments over random selection of molecules and so far these have been the most plentiful types of metrics that are routinely used to validate in silico models. The future of in silico pharmacology may be somewhat difficult to predict. While we are seeing a closer interaction between computational and in vitro approaches to date, will we see a similar relationship with in vivo studies in the future? More broadly, will in silico pharmacology ever be able to replace entirely experimental approaches in vitro and even in vivo, as some animal rights activists want us to believe? The answer here can only be a clear and resounding ‘no' (at least in the near future), for two irrefutable reasons. First, biological entities are nonlinear systems showing ‘chaotic behaviour'. As such, there is no relation between the magnitude of the input and the magnitude of the output, with even the most minuscule differences between initial conditions rapidly translating into major differences in the output. And second, no computer programme, however ‘complex and systems-like', will ever be able to fully model the complexity of biological systems. Indeed, and in the formulation of the mathematician Gregory Chaitin, biological systems are algorithmically incompressible, meaning that they cannot be modelled fully by an algorithm shorter than themselves.
In the meantime, in silico pharmacology will likely become more complex requiring some degree of integration of models, as we are seeing in the combined metabolism modelling approaches (Figure 3). Ultimately, to have a much broader impact, the in silico tools will need to become a part of every pharmacologist's tool kit and this will require training in modelling and informatics, alongside the in vivo, in vitro and molecular skills. This should provide a realistic appreciation of what the different in silico methods can and cannot be expected to do with regard to the pharmacologists aim of discovering new therapeutics.
Acknowledgments
SE gratefully acknowledges Dr Cheng Chang and Dr Peter W Swaan (University of Maryland), Dr Konstantin V Balakin (Chemical Diversity Inc.) for in silico pharmacology collaborations over the past several years and Dr Hugo Kubinyi for his insightful efforts in tabulating the successful applications of in silico approaches, which was inspirational. GoldenHelix Inc. graciously provided ChemTree. SE kindly acknowledges Dr Maggie AZ Hupcey for her support. JM acknowledges the research funding provided by the Spanish Ministerio de Educación y Ciencia (project reference BIO2005-04171) and the Instituto de Salud Carlos III. Owing to limited space, it was not possible to cite all in silico pharmacology-related papers, our sincere apologies to those omitted.
Abbreviations
- ADME/Tox
absorption, distribution, metabolism, excretion and toxicity
- CoMFA
comparative molecular field analysis
- COX
cyclooxygenase
- CYP
cytochrome P450
- EGFR
epidermal growth factor receptor
- ERα
oestrogen receptor subtype α
- hERG
human ether a-go-go related gene
- K-PLS
Kernel partial least squares
- kNN
k-nearest neighbours
- LOX
5 lipoxygenase
- MDDR
MDL Drug Data Reports
- MTree
multiple feature tree
- NCI
National Cancer Institute
- PCA
principal component analysis
- PD
pharmacodynamic
- PK
pharmacokinetic
- PLSs
partial least squares
- QSAR
quantitative structure–activity relationship
- RARα
retinoic acid receptor α
- SAR
structure–activity relationship
- SVM
support vector machine
Conflict of interest
The authors state no conflict of interest.
References
- Amin EA, Welsh WJ. A preliminary in silico lead series of 2-phthalimidinoglutaric acid analogues designed as MMP-3 inhibitors. J Chem Inf Model. 2006;46:2104–2109. doi: 10.1021/ci0601362. [DOI] [PubMed] [Google Scholar]
- Aronov AM, Balakin KV, Kiselyov A, Varma-O'Brien S, Ekins S.Applications of QSAR methods to ion channels Computational Toxicology: Risk Assessment for Pharmaceutical and Environmental Chemicals 2006John Wiley and Sons: Hoboken, NJ; In: Ekins S (ed)pp in press [Google Scholar]
- Balakin KV, Ekins S, Bugrim A, Ivanenkov YA, Korolev D, Nikolsky Y, et al. Quantitative structure–metabolism relationship modeling of the metabolic N-dealkylation rates. Drug Metab Dispos. 2004;32:1111–1120. doi: 10.1124/dmd.104.000364. [DOI] [PubMed] [Google Scholar]
- Balakin KV, Ivanenkov YA, Savchuk NP, Ivaschenko AA, Ekins S. Comprehensive computational assessment of ADME properties using mapping techniques. Curr Drug Disc Tech. 2005;2:99–113. doi: 10.2174/1570163054064666. [DOI] [PubMed] [Google Scholar]
- Baldwin ET, Bhat TN, Gulnick SV, Hosur MV, Sowder Il RC, Cachau RE, et al. Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA. 1993;90:6796–6800. doi: 10.1073/pnas.90.14.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreca ML, Gitto R, Quartarone S, De Luca L, De Sarro G, Chimirri A. Pharmacophore modeling as an efficient tool in the discovery of novel noncompetitive AMPA receptor antagonists. J Chem Inf Comput Sci. 2003;43:651–655. doi: 10.1021/ci025625q. [DOI] [PubMed] [Google Scholar]
- Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, et al. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J Med Chem. 2006;49:3116–3135. doi: 10.1021/jm0508641. [DOI] [PubMed] [Google Scholar]
- Bi X, Haque TS, Zhou J, Skillman AG, Lin B, Lee CE, et al. Novel cathepsin D inhibitors block the formation of hyperphosphorylated Tau fragments in hippocampus. J Neurochem. 2000;74:1469–1477. doi: 10.1046/j.1471-4159.2000.0741469.x. [DOI] [PubMed] [Google Scholar]
- Bonacci TM, Mathews JL, Yuan C, Lehmann DM, Malik S, Wu D, et al. Differential targeting of Gbetagamma-subunit signaling with small molecules. Science. 2006;312:443–446. doi: 10.1126/science.1120378. [DOI] [PubMed] [Google Scholar]
- Brea J, Rodrigo J, Carrieri A, Sanz F, Cadavid MI, Enguix MJ, et al. New serotonin 5-HT(2A), 5-HT(2B), and 5-HT(2C) receptor antagonists: synthesis, pharmacology, 3D-QSAR, and molecular modeling of (aminoalkyl)benzo and heterocycloalkanones. J Med Chem. 2002;45:54–71. doi: 10.1021/jm011014y. [DOI] [PubMed] [Google Scholar]
- Bursavich MG, Rich DH. Designing non-peptide peptidomimetics in the 21st century: inhibitors targeting conformational ensembles. J Med Chem. 2002;45:541–558. doi: 10.1021/jm010425b. [DOI] [PubMed] [Google Scholar]
- Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgenson WL, Lins RD, et al. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000;43:2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metab Dispos. 2006a;34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- Chang C, Ekins S, Bahadduri P, Swaan PW. Pharmacophore-based discovery of ligands for drug transporters. Adv Drug Del Rev. 2006b;58:1431–1450. doi: 10.1016/j.addr.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Swaan PW. Computational approaches to modeling drug transporters. Eur J Pharm Sci. 2005;27:411–424. doi: 10.1016/j.ejps.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Charlier C, Henichart JP, Durant F, Wouters J. Structural insights into human 5-lipoxygenase inhibition: combined ligand-based and target-based approach. J Med Chem. 2006;49:186–195. doi: 10.1021/jm050870x. [DOI] [PubMed] [Google Scholar]
- Chen D, Wang CY, Lambert JD, Ai N, Welsh WJ, Yang CS. Inhibition of human liver catechol-O-methyltransferase by tea catechins and their metabolites: structure–activity relationship and molecular-modeling studies. Biochem Pharmacol. 2005;69:1523–1531. doi: 10.1016/j.bcp.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Dajani R, Cleasby A, Neu M, Wonacott AJ, Jhoti H, Hood AM, et al. X-ray crystal structure of human dopamine sulfotransferase, SULT1A3. J Biol Chem. 1999;53:37862–37868. doi: 10.1074/jbc.274.53.37862. [DOI] [PubMed] [Google Scholar]
- de Graaf C, Vermeulen NP, Feenstra KA. Cytochrome P450 in silico: an integrative modeling approach. J Med Chem. 2005;48:2725–2755. doi: 10.1021/jm040180d. [DOI] [PubMed] [Google Scholar]
- de Groot MJ. Designing better drugs: predicting cytochrome P450 metabolism. Drug Discov Today. 2006;11:601–606. doi: 10.1016/j.drudis.2006.05.001. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Alex AA, Jones BC. Development of a combined protein and pharmacophore model for cytochrome P450 2C9. J Med Chem. 2002a;45:1983–1993. doi: 10.1021/jm0110791. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Ekins S. Pharmacophore modeling of cytochromes P450. Adv Drug Del Rev. 2002b;54:367–383. doi: 10.1016/s0169-409x(02)00009-1. [DOI] [PubMed] [Google Scholar]
- Debnath AK. Generation of predictive pharmacophore models for CCR5 antagonists: study with piperidine- and piperazine-based compounds as a new class of HIV-1 entry inhibitors. J Med Chem. 2003;46:4501–4515. doi: 10.1021/jm030265z. [DOI] [PubMed] [Google Scholar]
- Deng W, Breneman C, Embrechts MJ. Predicting protein-ligand binding affinities using novel geometrical descriptors and machine-learning methods. J Chem Inf Comput Sci. 2004;44:699–703. doi: 10.1021/ci034246+. [DOI] [PubMed] [Google Scholar]
- Di Santo R, Fermeglia M, Ferrone M, Paneni MS, Costi R, Artico M, et al. Simple but highly effective three-dimensional chemical-feature-based pharmacophore model for diketo acid derivatives as hepatitis C virus RNA-dependent RNA polymerase inhibitors. J Med Chem. 2005;48:6304–6314. doi: 10.1021/jm0504454. [DOI] [PubMed] [Google Scholar]
- Dimitrov S, Dimitrova G, Pavlov T, Dimitrova N, Patlewicz G, Niemela J, et al. A stepwise approach for defining the applicability domain of SAR and QSAR models. J Chem Inf Model. 2005;45:839–849. doi: 10.1021/ci0500381. [DOI] [PubMed] [Google Scholar]
- Doddareddy MR, Cho YS, Koh HY, Kim DH, Pae AN. In silico renal clearance model using classical Volsurf approach. J Chem Inf Model. 2006;46:1312–1320. doi: 10.1021/ci0503309. [DOI] [PubMed] [Google Scholar]
- Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, et al. Molecular docking and highthroughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- Ekins S, Balakin KV, Savchuk N, Ivanenkov Y. Insights for human ether-a-go-go-related gene potassium channel inhibition using recursive partitioning, Kohonen and Sammon mapping techniques. J Med Chem. 2006;49:5059–5071. doi: 10.1021/jm060076r. [DOI] [PubMed] [Google Scholar]
- Ekins S, Berbaum J, Harrison RK, Zecher M, Yuan J, Ishchenko AV, et al. Applying computational and in vitro approaches to lead selection Pharmaceutical Profiling in Drug Discovery for Lead Selection 2004aAAPS Press: Arlington, VA; 361–389.In: Borchardt RT, Kerns EH, Lipinski CA, Thakker DR, Wang B (eds). [Google Scholar]
- Ekins S, Bravi G, Binkley S, Gillespie JS, Ring BJ, Wikel JH, et al. Three and four dimensional-quantitative structure activity relationship analyses of CYP3A4 inhibitors. J Pharmacol Exp Ther. 1999a;290:429–438. [PubMed] [Google Scholar]
- Ekins S, Bravi G, Wikel JH, Wrighton SA. Three dimensional quantitative structure activity relationship (3D-QSAR) analysis of CYP3A4 substrates. J Pharmacol Exp Ther. 1999b;291:424–433. [PubMed] [Google Scholar]
- Ekins S, Mestres J, Testa B.In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling Br J Pharmacol 2007 10.1038/sj.bjp.0707305[E-pub ahead of print: 4 June 2007; doi] [DOI] [PMC free article] [PubMed]
- Ekins S, Nikolsky Y, Nikolskaya T. Techniques: application of systems biology to absorption, distribution, metabolism, excretion, and toxicity. Trends Pharmacol Sci. 2005;26:202–209. doi: 10.1016/j.tips.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Ekins S, Obach RS. Three dimensional-quantitative structure activity relationship computational approaches of prediction of human in vitro intrinsic clearance. J Pharmacol Exp Ther. 2000;295:463–473. [PubMed] [Google Scholar]
- Ekins S, Swaan PW. Development of computational models for enzymes, transporters, channels and receptors relevant to ADME/TOX. Rev Comp Chem. 2004b;20:333–415. [Google Scholar]
- Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer PM. The design of drug candidate molecules as selective inhibitors of therapeutically relevant protein kinases. Curr Med Chem. 2004;11:1563–1583. doi: 10.2174/0929867043365062. [DOI] [PubMed] [Google Scholar]
- Fisher NDL, Hollenberg NK. Is there a future for renin inhibitors. Expert Opin Invest Drugs. 2001;10:417–426. doi: 10.1517/13543784.10.3.417. [DOI] [PubMed] [Google Scholar]
- Fitzgerald LW, Burn TC, Brown BS, Patterson JP, Corjay MH, Valentine PA, et al. Possible role of valvular serotonin 5-HT(2B) receptors in the cardiopathy associated with fenfluramine. Mol Pharmacol. 2000;57:75–81. [PubMed] [Google Scholar]
- Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Analysis of drug-induced effect patterns to link structure and side effects of medicines. Nat Chem Biol. 2005a;1:389–397. doi: 10.1038/nchembio747. [DOI] [PubMed] [Google Scholar]
- Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biological spectra analysis: linking biological activity profiles to molecular structure. Proc Natl Acad Sci USA. 2005b;102:261–266. doi: 10.1073/pnas.0407790101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biospectra analysis: model proteome characterizations for linking molecular structure and biological response. J Med Chem. 2005c;48:6918–6925. doi: 10.1021/jm050494g. [DOI] [PubMed] [Google Scholar]
- Flohr S, Kurz M, Kostenis E, Brkovich A, Fournier A, Klabunde T. Identification of nonpeptidic urotensin II receptor antagonists by virtual screening based on a pharmacophore model derived from structure–activity relationships and nuclear magnetic resonance studies on urotensin II. J Med Chem. 2002;45:1799–1805. doi: 10.1021/jm0111043. [DOI] [PubMed] [Google Scholar]
- Fujita T. Recent success stories leading to commercializable bioactive compounds with the aid of traditional QSAR procedures. Quant Struct Act Relat. 1997;16:107–112. [Google Scholar]
- Fullbeck M, Huang X, Dumdey R, Frommel C, Dubiel W, Preissner R. Novel curcumin- and emodin-related compounds identified by in silico 2D/3D conformer screening induce apoptosis in tumor cells. BMC Cancer. 2005;5:97. doi: 10.1186/1471-2407-5-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet P, Zimmermann J, Capraro HG, Meyer T, Imbach P. Structure-based design of potent CDK1 inhibitors derived from olomoucine. J Comput Aided Mol Des. 2000;14:403–409. doi: 10.1023/a:1008115004986. [DOI] [PubMed] [Google Scholar]
- Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, et al. Structure of a human carcinogen-converting enzyme, SULT1A1. J Biol Chem. 2003;278:7655–7662. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- Ganguly M, Brown N, Schuffenhauer A, Ertl P, Gillet VJ, Greenidge PA. Introducing the consensus modeling concept in genetic algorithms: application to interpretable discriminant analysis. J Chem Inf Model. 2006;46:2110–2124. doi: 10.1021/ci050529l. [DOI] [PubMed] [Google Scholar]
- Ghafourian T, Rashidi MR. Quantitative study of the structural requirements of phthalazine/quinazoline derivatives for interaction with human liver aldehyde oxidase. Chem Pharm Bull (Tokyo) 2001;49:1066–1071. doi: 10.1248/cpb.49.1066. [DOI] [PubMed] [Google Scholar]
- Glen RC, Martin GR, Hill AP, Hyde RM, Woollard PM, Salmon JA, et al. Computer-aided design and synthesis of 5-substituted tryptamines and their pharmacology at the 5-HT1D receptor: discovery of compounds with potential anti-migraine properties. J Med Chem. 1995;38:3566–3580. doi: 10.1021/jm00018a016. [DOI] [PubMed] [Google Scholar]
- Gomeni R, Bani M, D'Angeli C, Corsi M, Bye A. Computer-assisted drug development (CADD): an emerging technology for designing first-time-in-man and proof-of-concept studies from preclinical experiments. Eur J Pharm Sci. 2001;13:261–270. doi: 10.1016/s0928-0987(01)00111-7. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan B, Aparna V, Jeevan J, Ravi M, Desiraju GR. A virtual screening approach for thymidine monophosphate kinase inhibitors as antitubercular agents based on docking and pharmacophore models. J Chem Inf Model. 2005;45:1101–1108. doi: 10.1021/ci050064z. [DOI] [PubMed] [Google Scholar]
- Griffith R, Luu TT, Garner J, Keller PA. Combining structure-based drug design and pharmacophores. J Mol Graph Model. 2005;23:439–446. doi: 10.1016/j.jmgm.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Guller R, Binggeli A, Breu D, Bur D, Fischli W, Hirth G, et al. Piperidine-renin inhibitors compounds with improved physicochemical properties. Bioorg Med Chem Lett. 1999;9:1403–1408. doi: 10.1016/s0960-894x(99)00196-1. [DOI] [PubMed] [Google Scholar]
- Guner O, Clement O, Kurogi Y. Pharmacophore modeling and three dimensional database searching for drug design using catalyst: recent advances. Curr Med Chem. 2004;11:2991–3005. doi: 10.2174/0929867043364036. [DOI] [PubMed] [Google Scholar]
- Hancock CN, Macias A, Lee EK, Yu SY, Mackerell AD, Jr, Shapiro P. Identification of novel extracellular signal-regulated kinase docking domain inhibitors. J Med Chem. 2005;48:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- Hansch C, Lien EJ, Helmer F. Structure–activity correlations in the metabolism of drugs. Arch Biochem Biophys. 1968;128:319–330. doi: 10.1016/0003-9861(68)90038-6. [DOI] [PubMed] [Google Scholar]
- Hert J, Willett P, Wilton DJ, Acklin P, Azzaoui K, Jacoby E, et al. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J Chem Inf Comput Sci. 2004;44:1177–1185. doi: 10.1021/ci034231b. [DOI] [PubMed] [Google Scholar]
- Hessler G, Zimmermann M, Matter H, Evers A, Naumann T, Lengauer T, et al. Multiple-ligand-based virtual screening: methods and applications of the MTree approach. J Med Chem. 2005;48:6575–6584. doi: 10.1021/jm050078w. [DOI] [PubMed] [Google Scholar]
- Hibert MF, Gittos MW, Middlemiss DN, Mir AK, Fozard JR. Graphics computer-aided receptor mapping as a predictive tool for drug design: development of potent, selective, and stereospecific ligands for the 5-HT1A receptor. J Med Chem. 1988;31:1087–1093. doi: 10.1021/jm00401a007. [DOI] [PubMed] [Google Scholar]
- Hirashima A, Tomita J, Pan C, Taniguchi E, Eto M. Quantitative structure-activity studies of octopaminergic 2-(arylimino)thiazolidines and oxazolidines against the nervous system of Periplaneta americana L. Bioorg Med Chem. 1997;5:2121–2128. doi: 10.1016/s0968-0896(97)00136-3. [DOI] [PubMed] [Google Scholar]
- Hirst WD, Abrahamsen B, Blaney FE, Calver AR, Aloj L, Price GW, et al. Differences in the central nervous system distribution and pharmacology of the mouse 5-hydroxytryptamine-6 receptor compared with rat and human receptors investigated by radioligand binding, site-directed mutagenesis, and molecular modeling. Mol Pharmacol. 2003;64:1295–1308. doi: 10.1124/mol.64.6.1295. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Huang N, Nagarsekar A, Xia G, Hayashi J, MacKerell AD., Jr Identification of non-phosphate-containing small molecular weight inhibitors of the tyrosine kinase p56 Lck SH2 domain via in silico screening against the pY + 3 binding site. J Med Chem. 2004;47:3502–3511. doi: 10.1021/jm030470e. [DOI] [PubMed] [Google Scholar]
- Huo S, Wang J, Cieplak P, Kollman PA, Kuntz ID. Molecular dynamics and free energy analyses of cathepsin D-inhibitor interactions: insight into structure-based ligand design. J Med Chem. 2002;45:1412–1419. doi: 10.1021/jm010338j. [DOI] [PubMed] [Google Scholar]
- Ji ZL, Wang Y, Yu L, Han LY, Zheng CJ, Chen YZ. In silico search of putative adverse drug reaction related proteins as a potential tool for facilitating drug adverse effect prediction. Toxicol Lett. 2006;164:104–112. doi: 10.1016/j.toxlet.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Kaminski JJ, Rane DF, Snow ME, Weber L, Rothofsky ML, Anderson SD, et al. Identification of novel farnesyl protein transferase inhibitors using three-dimensional searching methods. J Med Chem. 1997;40:4103–4112. doi: 10.1021/jm970291v. [DOI] [PubMed] [Google Scholar]
- Kang J, Wang L, Chen X-L, Triggle DJ, Rampe D. Interactions of a series of fluoroquinolone antibacterial drugs with the human cardiac K+ channel HERG. Mol Pharmacol. 2001;59:122–126. doi: 10.1124/mol.59.1.122. [DOI] [PubMed] [Google Scholar]
- Kansal AR. Modeling approaches to type 2 diabetes. Diabetes Technol Ther. 2004;6:39–47. doi: 10.1089/152091504322783396. [DOI] [PubMed] [Google Scholar]
- Kauvar LM, Higgins DL, Villar HO, Sportsman JR, Engqvist-Goldstein A, Bukar R, et al. Predicting ligand binding to proteins by affinity fingerprinting. Chem Biol. 1995;2:107–118. doi: 10.1016/1074-5521(95)90283-x. [DOI] [PubMed] [Google Scholar]
- Kauvar LM, Laborde E. The diversity challenge in combinatorial chemistry. Curr Opin Drug Disc Dev. 1998a;1:66–70. [PubMed] [Google Scholar]
- Kauvar LM, Villar HO, Sportsman JR, Higgins DL, Schmidt DEJ. Protein affinity map of chemical space. J Chromatography B. 1998b;715:93–102. doi: 10.1016/s0378-4347(98)00045-0. [DOI] [PubMed] [Google Scholar]
- Keenan SM, DeLisle RK, Welsh WJ, Paula S, Ball WJ., Jr Elucidation of the Na+, K+-ATPase digitalis binding site. J Mol Graph Model. 2005;23:465–475. doi: 10.1016/j.jmgm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Kenyon V, Chorny I, Carvajal WJ, Holman TR, Jacobson MP. Novel human lipoxygenase inhibitors discovered using virtual screening with homology models. J Med Chem. 2006;49:1356–1363. doi: 10.1021/jm050639j. [DOI] [PubMed] [Google Scholar]
- Khadikar P, Jaiswal M, Gupta M, Mandloi D, Sisodia RS. QSAR studies on 1,2-dithiole-3-thiones: modeling of lipophilicity, quinone reductase specific activity, and production of growth hormone. Bioorg Med Chem Lett. 2005;15:1249–1255. doi: 10.1016/j.bmcl.2004.11.074. [DOI] [PubMed] [Google Scholar]
- Kick EK, Roe DC, Skillman AG, Liu G, Ewing TJ, Sun Y, et al. Structure-based design and combinatorial chemistry yield low nanomolar inhibitors of cathepsin D. Chem Biol. 1997;4:297–307. doi: 10.1016/s1074-5521(97)90073-9. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Wendt KU, Kadereit D, Brachvogel V, Burger HJ, Herling AW, et al. Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. J Med Chem. 2005;48:6178–6193. doi: 10.1021/jm049034y. [DOI] [PubMed] [Google Scholar]
- Koide Y, Hasegawa T, Takahashi A, Endo A, Mochizuki N, Nakagawa M, et al. Development of novel EDG3 antagonists using a 3D database search and their structure–activity relationships. J Med Chem. 2002;45:4629–4638. doi: 10.1021/jm020080c. [DOI] [PubMed] [Google Scholar]
- Krovat EM, Langer T. Impact of scoring functions on enrichment in docking-based virtual screening: an application study on renin inhibitors. J Chem Inf Comput Sci. 2004;44:1123–1129. doi: 10.1021/ci0342728. [DOI] [PubMed] [Google Scholar]
- Kubinyi H.Success stories of computer-aided design Computer Applications in Pharmaceutical Research and Development 2006John Wiley and Sons: Hoboken; 377–424.In: Ekins S (ed) [Google Scholar]
- Kunick C, Zeng Z, Gussio R, Zaharevitz D, Leost M, Totzke F, et al. Structure-aided optimization of kinase inhibitors derived from alsterpaullone. Chembiochem. 2005;6:541–549. doi: 10.1002/cbic.200400099. [DOI] [PubMed] [Google Scholar]
- Kurogi Y, Guner OF. Pharmacophore modeling and three-dimensional database searching for drug design using catalyst. Curr Med Chem. 2001a;8:1035–1055. doi: 10.2174/0929867013372481. [DOI] [PubMed] [Google Scholar]
- Kurogi Y, Miyata K, Okamura T, Hashimoto K, Tsutsumi K, Nasu M, et al. Discovery of novel mesangial cell proliferation inhibitors using a three-dimensional database searching method. J Med Chem. 2001b;44:2304–2307. doi: 10.1021/jm010060v. [DOI] [PubMed] [Google Scholar]
- Langer T, Eder M, Hoffmann RD, Chiba P, Ecker GF. Lead identification for modulators of multidrug resistance based on in silico screening with a pharmacophoric feature model. Arch Pharm (Weinheim) 2004;337:317–327. doi: 10.1002/ardp.200300817. [DOI] [PubMed] [Google Scholar]
- Lepp Z, Kinoshita T, Chuman H. Screening for new antidepressant leads of multiple activities by support vector machines. J Chem Inf Model. 2006;46:158–167. doi: 10.1021/ci050301y. [DOI] [PubMed] [Google Scholar]
- Leung D, Abbenante G, Fairlie DP. Protease inhibitors: current status and future prospects. J Med Chem. 2000;43:305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- Lewis DFV. On the recognition of mammalian microsomal cytochrome P450 substrates and their characteristics. Biochem Pharmacol. 2000;60:293–306. doi: 10.1016/s0006-2952(00)00335-x. [DOI] [PubMed] [Google Scholar]
- Lill MA, Dobler M, Vedani A. Prediction of small-molecule binding to cytochrome P450 3A4: flexible docking combined with multidimensional QSAR. ChemMedChem. 2006;6:73–81. doi: 10.1002/cmdc.200500024. [DOI] [PubMed] [Google Scholar]
- Liu Z, Huang C, Fan K, Wei P, Chen H, Liu S, et al. Virtual screening of novel noncovalent inhibitors for SARS-CoV 3C-like proteinase. J Chem Inf Model. 2005;45:10–17. doi: 10.1021/ci049809b. [DOI] [PubMed] [Google Scholar]
- Lloyd DG, Golfis G, Knox AJS, Fayne D, Meegan MJ, Oprea TI. Oncology exploration: chartering cancer medicinal chemistry space. DDT. 2006;11:149–159. doi: 10.1016/S1359-6446(05)03688-3. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Obach RS, Dicapua FM, Bakken GA, Lu J, Potter DM, et al. A hybrid mixture discriminant analysis-random forest computational model for the prediction of volume of distribution of drugs in human. J Med Chem. 2006;49:2262–2267. doi: 10.1021/jm050200r. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Obach RS, Shalaeva MY, Gao F. Prediction of human volume of distribution values for neutral and basic drugs. 2. Extended data set and leave-class-out statistics. J Med Chem. 2004;47:1242–1250. doi: 10.1021/jm030408h. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Obach RS, Shalaeva MY, Gao F. Prediction of volume of distribution values in humans for neutral and basic drugs using physicochemical measurements and plasma protein binding. J Med Chem. 2002;45:2867–2876. doi: 10.1021/jm0200409. [DOI] [PubMed] [Google Scholar]
- Lorenzen S, Dunkel M, Preissner R. In silico screening of drug databases for TSE inhibitors. Biosystems. 2005;80:117–122. doi: 10.1016/j.biosystems.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Lu IL, Huang CF, Peng YH, Lin YT, Hsieh HP, Chen CT, et al. Structure-based drug design of a novel family of PPARgamma partial agonists: virtual screening, X-ray crystallography, and in vitro/in vivo biological activities. J Med Chem. 2006;49:2703–2712. doi: 10.1021/jm051129s. [DOI] [PubMed] [Google Scholar]
- Mager H, Schneeweiss B, Barth A, Mager PP. Judging models in QSAR- and LFE-like studies if there are no replications: correlation of dipeptidyl peptidase IV hydrolytic activities of L-alanyl-L-alanine phenylamides. Pharmazie. 1982;37:856–857. [PubMed] [Google Scholar]
- Manivet P, Schneider B, Smith JC, Choi DS, Maroteaux L, Kellermann O, et al. The serotonin binding site of human and murine 5-HT2B receptors: molecular modeling and site-directed mutagenesis. J Biol Chem. 2002;277:17170–17178. doi: 10.1074/jbc.M200195200. [DOI] [PubMed] [Google Scholar]
- Mao B, Gozalbes R, Barbosa F, Migeon J, Merrick S, Kamm K, et al. QSAR modeling of in vitro inhibition of cytochrome P450 3A4. J Chem Inf Model. 2006;46:2125–2134. doi: 10.1021/ci0600915. [DOI] [PubMed] [Google Scholar]
- Marki HP, Binggeli A, Bittner B, Bohner-Lang V, Breu V, Bur D, et al. Piperidine renin inhibitors: from leads to drugs. Il Farmaco. 2001;56:21–27. doi: 10.1016/s0014-827x(01)01004-7. [DOI] [PubMed] [Google Scholar]
- McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- McGovern SL, Shoichet BK. Kinase inhibitors: not just for kinases anymore. J Med Chem. 2003;46:1478–1483. doi: 10.1021/jm020427b. [DOI] [PubMed] [Google Scholar]
- Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov Today. 2004;9:641–651. doi: 10.1016/S1359-6446(04)03163-0. [DOI] [PubMed] [Google Scholar]
- Mullally JE, Fitzpatrick FA. Pharmacophore model for novel inhibitors of ubiquitin isopeptidases that induce p53-independent cell death. Mol Pharmacol. 2002;62:351–358. doi: 10.1124/mol.62.2.351. [DOI] [PubMed] [Google Scholar]
- Mullally JE, Moos PJ, Edes K, Fitzpatrick FA. Cyclopentenone prostaglandins of the J series inhibit the ubiquitin isopeptidase activity of the proteasome pathway. J Biol Chem. 2001;276:30366–30373. doi: 10.1074/jbc.M102198200. [DOI] [PubMed] [Google Scholar]
- Nebigil CG, Choi DS, Dierich A, Hickel P, Le Meur M, Messaddeq N, et al. Serotonin 2B receptor is required for heart development. Proc Natl Acad Sci USA. 2000;97:9508–9513. doi: 10.1073/pnas.97.17.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklaus MC, Neamati N, Hong H, Mazumder A, Sunder S, Chen J, et al. HIV-1 integrase pharmacophore: discovery of inhibitors through three-dimensional database searching. J Med Chem. 1997;40:920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- Niimi T, Orita M, Okazawa-Igarashi M, Sakashita H, Kikuchi K, Ball E, et al. Design and synthesis of non-peptidic inhibitors for the Syk C-terminal SH2 domain based on structure-based in-silico screening. J Med Chem. 2001;44:4737–4740. doi: 10.1021/jm010313k. [DOI] [PubMed] [Google Scholar]
- Niwa T. Prediction of biological targets using probabilistic neural networks and atom-type descriptors. J Med Chem. 2004;47:2645–2650. doi: 10.1021/jm0302795. [DOI] [PubMed] [Google Scholar]
- Noble D, Colatsky TJ. A return to rational drug discovery: computer-based models of cells, organs and systems in drug target identification. Emerging therapeutic targets. 2000;4:39–49. [Google Scholar]
- Nowak M, Kolaczkowski M, Pawlowski M, Bojarski AJ. Homology modeling of the serotonin 5-HT1A receptor using automated docking of bioactive compounds with defined geometry. J Med Chem. 2006;49:205–214. doi: 10.1021/jm050826h. [DOI] [PubMed] [Google Scholar]
- O'Brien SE, Brown DG, Mills JE, Phillips C, Morris G. Computational tools for the analysis and visualization of multiple protein–ligand complexes. J Mol Graph Model. 2005;24:186–194. doi: 10.1016/j.jmgm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Oefner C, Binggeli A, Breu D, Bur D, Clozel J-P, D'Arcy A, et al. Renin inhibition by substituted piperidines: a novel paradigm for the inhibition of monomeric aspartic proteinases. Chem Biol. 1999;6:127–131. doi: 10.1016/s1074-5521(99)89004-8. [DOI] [PubMed] [Google Scholar]
- Oloff S, Mailman RB, Tropsha A. Application of validated QSAR models of D1 dopaminergic antagonists for database mining. J Med Chem. 2005;48:7322–7332. doi: 10.1021/jm049116m. [DOI] [PubMed] [Google Scholar]
- Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- Pegg SC-H, Haresco JJ, Kuntz ID. A genetic algorithm for structure-based de novo design. J Comput Aided Mol Des. 2001;15:911–933. doi: 10.1023/a:1014389729000. [DOI] [PubMed] [Google Scholar]
- Peng T, Pei J, Zhou J. 3D-QSAR and receptor modeling of tyrosine kinase inhibitors with flexible atom receptor model (FLARM) J Chem Inf Comput Sci. 2003;43:298–303. doi: 10.1021/ci0256034. [DOI] [PubMed] [Google Scholar]
- Pleban K, Kaiser D, Kopp S, Peer M, Chiba P, Ecker GF. Targeting drug-efflux pumps – a pharmacoinformatic approach. Acta Biochim Pol. 2005;52:737–740. [PubMed] [Google Scholar]
- Rahuel J, Rasetti V, Maibaum J, Rueger H, Goschke R, Cohen N-C, et al. Structure-based drug design: the discovery of novel nonpeptide orally active inhibitors of human renin. Chem Biol. 2000;7:493–504. doi: 10.1016/s1074-5521(00)00134-4. [DOI] [PubMed] [Google Scholar]
- Rollinger JM, Bodensieck A, Seger C, Ellmerer EP, Bauer R, Langer T, et al. Discovering COX-inhibiting constituents of Morus root bark: activity-guided versus computer-aided methods. Planta Med. 2005;71:399–405. doi: 10.1055/s-2005-864132. [DOI] [PubMed] [Google Scholar]
- Rollinger JM, Hornick A, Langer T, Stuppner H, Prast H. Acetylcholinesterase inhibitory activity of scopolin and scopoletin discovered by virtual screening of natural products. J Med Chem. 2004;47:6248–6254. doi: 10.1021/jm049655r. [DOI] [PubMed] [Google Scholar]
- Roth BL, Lopez E, Beischel S, Westkaemper RB, Evans JM. Screening the receptorome to discover the molecular targets for plant-derived psychoactive compounds: a novel approach for CNS drug discovery. Pharmacol Ther. 2004;102:99–110. doi: 10.1016/j.pharmthera.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Rothman RB, Baumann MH, Savage JE, Rauser L, McBride A, Hufeisen SJ, et al. Evidence for possible involvement of 5-HT(2B) receptors in the cardiac valvulopathy associated with fenfluramine and other serotonergic medications. Circulation. 2000;102:2836–2841. doi: 10.1161/01.cir.102.23.2836. [DOI] [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- Schapira M, Raaka BM, Samuels HH, Abagyan R. Rational discovery of novel nuclear hormone receptor antagonists. Proc Natl Acad Sci USA. 2000;97:1008–1013. doi: 10.1073/pnas.97.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Coassolo P, Lave T. Combining in vitro and in vivo pharmacokinetic data for prediction of hepatic drug clearance in humans by artificial neural networks and multivariate statistical techniques. J Med Chem. 1999;42:5072–5076. doi: 10.1021/jm991030j. [DOI] [PubMed] [Google Scholar]
- Schuster D, Laggner C, Steindl TM, Palusczak A, Hartmann RW, Langer T. Pharmacophore modeling and in silico screening for new P450 19 (aromatase) inhibitors. J Chem Inf Model. 2006;46:1301–1311. doi: 10.1021/ci050237k. [DOI] [PubMed] [Google Scholar]
- Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- Setola V, Dukat M, Glennon RA, Roth BL. Molecular determinants for the interaction of the valvulopathic anorexigen norfenfluramine with the 5-HT2B receptor. Mol Pharmacol. 2005;68:20–33. doi: 10.1124/mol.104.009266. [DOI] [PubMed] [Google Scholar]
- Shao D, Berrodin TJ, Manas E, Hauze D, Powers R, Bapat A, et al. Identification of novel estrogen receptor alpha antagonists. J Steroid Biochem Mol Biol. 2004;88:351–360. doi: 10.1016/j.jsbmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Sharom JR, Bellows DS, Tyers M. From large networks to small molecules. Curr Opin Chem Biol. 2004;8:81–90. doi: 10.1016/j.cbpa.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Sheridan RP, Feuston BP, Maiorov VN, Kearsley SK. Similarity to molecules in the training set is a good discriminator for prediction accuracy in QSAR. J Chem Inf Comput Sci. 2004;44:1912–1928. doi: 10.1021/ci049782w. [DOI] [PubMed] [Google Scholar]
- Shi Y. Orphan nuclear receptors, excellent targets of drug discovery. Comb Chem High Throughput Screen. 2006;9:683–689. doi: 10.2174/138620706778700125. [DOI] [PubMed] [Google Scholar]
- Shimada J.The challenges of making useful protein–ligand free energy predictions for drug discovery Computer Applications in Pharmaceutical Research and Development 2006John Wiley and Sons: Hoboken; 321–351.In: Ekins S (ed) [Google Scholar]
- Singh J, van Vlijmen H, Lee WC, Liao Y, Lin KC, Ateeq H, et al. 3D QSAR (COMFA) of a series of potent and highly selective VLA-4 antagonists. J Comput Aided Mol Des. 2002a;16:201–211. doi: 10.1023/a:1020130418084. [DOI] [PubMed] [Google Scholar]
- Singh J, Van Vlijmen H, Liao Y, Lee WC, Cornebise M, Harris M, et al. Identification of potent and novel alpha4beta1 antagonists using in silico screening. J Med Chem. 2002b;45:2988–2993. doi: 10.1021/jm020054e. [DOI] [PubMed] [Google Scholar]
- Singh P, Kumar R. Quantitative structure–activity relationship study on tetrahydro-beta-carboline antagonists of the serotonin 2B (5HT2B) contractile receptor in the rat stomach fundus. J Enzyme Inhib. 2001;16:491–497. doi: 10.1080/14756360127570. [DOI] [PubMed] [Google Scholar]
- Sipila J, Taskinen J. CoMFA modeling of human catechol O-methyltransferase enzyme kinetics. J Chem Inf Comput Sci. 2004;44:97–104. doi: 10.1021/ci034189k. [DOI] [PubMed] [Google Scholar]
- Sippl W. Development of biologically active compounds by combining 3D QSAR and structure-based design methods. J Comput Aided Mol Des. 2002;16:825–830. doi: 10.1023/a:1023888813526. [DOI] [PubMed] [Google Scholar]
- Smith PA, Sorich MJ, Low LS, McKinnon RA, Miners JO. Towards integrated ADME prediction: past, present and future directions for modelling metabolism by UDP-glucuronosyltransferases. J Mol Graph Model. 2004;22:507–517. doi: 10.1016/j.jmgm.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Sorich MJ, Miners JO, McKinnon RA, Smith PA. Multiple pharmacophores for the investigation of human UDP-glucuronosyltransferase isoform substrate selectivity. Mol Pharmacol. 2004;65:301–308. doi: 10.1124/mol.65.2.301. [DOI] [PubMed] [Google Scholar]
- Srinivasen J, Castellino A, Bradley E, Eksterowicz JE, Grootenhuis PDJ, Putta S, et al. Evaluation of a novel shape-based computational filter for lead evolution: application to thrombin inhibitors. J Med Chem. 2002;45:2494–2500. doi: 10.1021/jm010494q. [DOI] [PubMed] [Google Scholar]
- Stanton A. Potential of renin inhibition in cardiovascular disease. JRAAS. 2003;4:6–10. doi: 10.3317/jraas.2003.008. [DOI] [PubMed] [Google Scholar]
- Steindl T, Laggner C, Langer T. Human rhinovirus 3C protease: generation of pharmacophore models for peptidic and nonpeptidic inhibitors and their application in virtual screening. J Chem Inf Model. 2005a;45:716–724. doi: 10.1021/ci049638a. [DOI] [PubMed] [Google Scholar]
- Steindl T, Langer T. Influenza virus neuraminidase inhibitors: generation and comparison of structure-based and common feature pharmacophore hypotheses and their application in virtual screening. J Chem Inf Comput Sci. 2004;44:1849–1856. doi: 10.1021/ci049844i. [DOI] [PubMed] [Google Scholar]
- Steindl TM, Crump CE, Hayden FG, Langer T. Pharmacophore modeling, docking, and principal component analysis based clustering: combined computer-assisted approaches to identify new inhibitors of the human rhinovirus coat protein. J Med Chem. 2005b;48:6250–6260. doi: 10.1021/jm050343d. [DOI] [PubMed] [Google Scholar]
- Strachan RT, Ferrara G, Roth BL. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov Today. 2006;11:708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Swaan PW, Ekins S. Reengineering the pharmaceutical industry by crash-testing molecules. Drug Disc Today. 2005;10:1191–1200. doi: 10.1016/S1359-6446(05)03557-9. [DOI] [PubMed] [Google Scholar]
- Terlau H, Stuhmer W. Structure and function of voltage-gated ion channels. Naturwissenschaften. 1998;85:437–444. doi: 10.1007/s001140050527. [DOI] [PubMed] [Google Scholar]
- Tervo AJ, Kyrylenko S, Niskanen P, Salminen A, Leppanen J, Nyronen TH, et al. An in silico approach to discovering novel inhibitors of human sirtuin type 2. J Med Chem. 2004;47:6292–6298. doi: 10.1021/jm049933m. [DOI] [PubMed] [Google Scholar]
- Tesmer JJG. Hitting the hot spots of cell signaling cascades. Science. 2006;312:377–378. doi: 10.1126/science.1126903. [DOI] [PubMed] [Google Scholar]
- Testa B, Krämer SD. The biochemistry of drug metabolism – an introduction. Part 1: principles and overview. Chem Biodevers. 2006;3:1053–1101. doi: 10.1002/cbdv.200690111. [DOI] [PubMed] [Google Scholar]
- Tetko IV, Bruneau P, Mewes HW, Rohrer DC, Poda GI. Can we estimate the accuracy of ADME-Tox predictions. Drug Discov Today. 2006;11:700–707. doi: 10.1016/j.drudis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Torres AM, Bansal P, Alewood PF, Bursill JA, Kuchel PW, Vandenberg JI. Solution structure of CnErg1 (ergtoxin), a HERG specific scorpion toxin. FEBS Lett. 2003;539:138–142. doi: 10.1016/s0014-5793(03)00216-3. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Chaki H, Takakura T, Kotsubo H, Tanaka T, Aikawa Y, et al. Discovery of nonpeptidic small-molecule AP-1 inhibitors: lead hopping based on a three-dimensional pharmacophore model. J Med Chem. 2006;49:80–91. doi: 10.1021/jm050550d. [DOI] [PubMed] [Google Scholar]
- Van Drie JH. Protein-structure-based drug discovery of renin inhibitors. BioCad user notes. 1993.
- Verbitski SM, Mullally JE, Fitzpatrick FA, Ireland CM. Punaglandins, chlorinated prostaglandins, function as potent Michael receptors to inhibit ubiquitin isopeptidase activity. J Med Chem. 2004;47:2062–2070. doi: 10.1021/jm030448l. [DOI] [PubMed] [Google Scholar]
- Vieira E, Binggeli A, Breu D, Bur D, Fischli W, Guller R, et al. Substituted piperidines-highly potent renin inhibitors due to induced fit adaption of the active site. Bioorg Med Chem Lett. 1999;9:1397–1402. doi: 10.1016/s0960-894x(99)00195-x. [DOI] [PubMed] [Google Scholar]
- Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Kinomics-structural biology and chemogenomics of kinase inhibitors and targets. Biochim Biophys Acta. 2004;1697:243–257. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Waller CL, Evans MV, McKinney JD. Modeling the cytochrome P450-mediated metabolism of chlorinated volatile organic compounds. Drug Metab Dispos. 1996;24:203–210. [PubMed] [Google Scholar]
- Wang S, Zaharevitz DW, Sharma R, Marquez VE, Lewin NE, Du L, et al. The discovery of novel, structurally diverse protein kinase C agonists through computer 3D-database pharmacophore search. Molecular modeling studies. J Med Chem. 1994;37:4479–4489. doi: 10.1021/jm00052a007. [DOI] [PubMed] [Google Scholar]
- Wielert-Badt S, Lin JT, Lorenz M, Fritz S, Kinne RK. Probing the conformation of the sugar transport inhibitor phlorizin by 2D-NMR, molecular dynamics studies, and pharmacophore analysis. J Med Chem. 2000;43:1692–1698. doi: 10.1021/jm9905460. [DOI] [PubMed] [Google Scholar]
- Willett P. Similarity-based approaches to virtual screening. Biochem Soc Trans. 2003;31:603–606. doi: 10.1042/bst0310603. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- Wong BR, Parlati F, Qu K, Demo S, Pray T, Huang J, et al. Drug discovery in the ubiquitin regulatory pathway. Drug Discov Today. 2003;8:746–754. doi: 10.1016/s1359-6446(03)02780-6. [DOI] [PubMed] [Google Scholar]
- Yu J, Paine MJ, Marechal JD, Kemp CA, Ward CJ, Brown S, et al. In silico prediction of drug binding to CYP2D6: identification of a new metabolite of metoclopramide. Drug Metab Dispos. 2006;34:1386–1392. doi: 10.1124/dmd.106.009852. [DOI] [PubMed] [Google Scholar]
- Zhang EY, Knipp GT, Ekins S, Swaan PW. Structural biology and function of solute transporters: implications for identifying and designing substrates. Drug Metab Rev. 2002a;34:709–750. doi: 10.1081/dmr-120015692. [DOI] [PubMed] [Google Scholar]
- Zhang EY, Phelps MA, Cheng C, Ekins S, Swaan PW. Modeling of active transport systems. Adv Drug Del Rev. 2002b;54:329–354. doi: 10.1016/s0169-409x(02)00007-8. [DOI] [PubMed] [Google Scholar]
- Zhao L, Brinton RD. Structure-based virtual screening for plant-based ERbeta-selective ligands as potential preventative therapy against age-related neurodegenerative diseases. J Med Chem. 2005;48:3463–3466. doi: 10.1021/jm0490538. [DOI] [PubMed] [Google Scholar]