Abstract
Study Objective:
A close association between the human leukocyte antigen (HLA)-DRB1*1501/DQB1*0602 and abnormalities in some inflammatory cytokines have been demonstrated in narcolepsy. Specific alterations in the immune system have been suggested to occur in this disorder. We attempted to identify alterations in gene expression underlying the abnormalities in the blood cells of narcoleptic patients.
Designs:
Total RNA from 12 narcolepsy-cataplexy patients and from 12 age- and sex-matched healthy controls were pooled. The pooled samples were initially screened for candidate genes for narcolepsy by differential display analysis using annealing control primers (ACP). The second screening of the samples was carried out by semiquantitative PCR using gene-specific primers. Finally, the expression levels of the candidate genes were further confirmed by quantitative real-time PCR using a new set of samples (20 narcolepsy-cataplexy patients and 20 healthy controls).
Results:
The second screening revealed differential expression of 4 candidate genes. Among them, MX2 was confirmed as a significantly down-regulated gene in the white blood cells of narcoleptic patients by quantitative real-time PCR.
Conclusion:
We found the MX2 gene to be significantly less expressed in comparison with normal subjects in the white blood cells of narcoleptic patients. This gene is relevant to the immune system. Although differential display analysis using ACP technology has a limitation in that it does not help in determining the functional mechanism underlying sleep/wakefulness dysregulation, it is useful for identifying novel genetic factors related to narcolepsy, such as HLA molecules. Further studies are required to explore the functional relationship between the MX2 gene and narcolepsy pathophysiology.
Citation:
Tanaka S; Honda Y; Honda M. Identification of differentially expressed genes in blood cells of narcolepsy patients. SLEEP 2007;30(8):974-979.
Keywords: Narcolepsy, differential display, blood cell, MX2
INTRODUCTION
NARCOLEPSY IS A DISABLING DISORDER CHARACTERIZED BY RECURRENT DAYTIME SLEEPINESS AND CATAPLEXY, WITH OR WITHOUT HYPNAGOGIC HALLUCINATIONS, and sleep paralysis.1 An immunological pathogenesis for narcolepsy has been suggested in view of the close association between narcolepsy with cataplexy and the human leukocyte antigen (HLA)-DRB1*1501-DQB1*0602. However, it remains to be clarified whether any autoimmune mechanism or immune-related alteration is involved in the pathophysiology of narcolepsy.2
There is evidence that the inflammatory cytokines tumor necrosis factor (TNF)-alpha and interleukin (IL)-6 are involved in physiological sleep regulation and are associated with sleepiness.3 The levels of the TNF-alpha subtypes and IL-6 are significantly elevated in narcolepsy patients as compared to those in normal controls.4 A significant association between a single nucleotide polymorphism in the TNF-alpha gene promoter or in the TNF receptor 2 gene and human narcolepsy was suggested.5,6 Himmerich et al demonstrated increased plasma levels of soluble TNF receptor p75, which is one of the TNF receptors in serum, in patients with narcolepsy having cataplexy.7 A functional alteration of the TNF-alpha cytokine system is suggested. Intravenous immunoglobulin that might help in suppressing proinflammatory cytokines reduced the number of cataplexy attacks in narcoleptic patients at disease onset.8 Recently, it was found that the level of C-reactive protein, an inflammatory-associated factor, was increased in the serum of hypocretin-1 deficient narcoleptic patients9 as compared to that in healthy controls after body mass index adjustment.
We hypothesized that some immunological dysregulations occur in narcoleptic patients that are reflected in the gene expression changes in white blood cells. Genes expressed in white blood cells are not limited to but are often related to the immune mechanism. In this study, we attempted to identify differentially expressed genes in the white blood cells of patients with narcolepsy with cataplexy by a differential display method using annealing control primers (ACPs). Our goal was to identify biological markers of narcolepsy in white blood cells.
METHODS
This research was approved by the ethical committees of the collaborative institutes. Written informed consents were obtained from all participants. All patients were diagnosed clinically as having narcolepsy with cataplexy according to the International Classification of Sleep Disorders, 2nd edition (ICSD-2)1 at the Neuropsychiatric Research Institute (Tokyo, Japan). All narcoleptic patients were HLA-DRB1*1501/DQB1*0602 positive. Blood samples and clinical data were collected at the Neuropsychiatric Research Institute and the Tokyo Institute of Psychiatry (Tokyo, Japan). None of the control subjects experienced excessive daytime sleepiness or exhibit any signs of immunological abnormalities based on the questionnaires obtained at the time of blood collection. HLA typing for the HLA-DRB1 and DQB1 loci was performed for all the subjects at the NPO HLA Laboratory (Kyoto, Japan). All 32 narcoleptic patients and 7 of the 32 control subjects (4 of the 12 subjects used for initial screening and 3 of the 20 subjects used in the validation study) tested positive for HLA-DRB1*1501-DQB1*0602.
Five milliliters of whole blood was collected in PAXgene Blood RNA tubes (Becton Dickinson, UK). Total RNA was isolated using a PAXgene Blood RNA Kit (QIAGEN, CA), according to the manufacturer's protocol. The eluted total RNA was treated with DNase I and cleaned using an RNeasy MinElute Cleanup kit (QIAGEN) to obtain concentrated total RNA. For the first screening, total RNA from 12 narcolepsy patients with a disease onset of 10 years or less and total RNA from age- and sex-matched 12 healthy controls were pooled. We used pooled samples to reduce individual variation and enhance the gene expression changes specific for narcolepsy. To compare the differentially expressed genes in the white blood cells of the narcolepsy patients and the healthy control patients, GeneFishing DEG kits (Seegene, Korea) were used according to the manufacturer's instructions. The synthesis of single-stranded cDNA was carried out using 3 μg of the total RNA from each pool, ReverTra Ace (TOYOBO, Japan), and dT-ACP1 whose 3′-end core portion comprises a hybridizing sequence complementary to the poly-A region of the mRNA transcripts. In brief, PCR was conducted by using arbitrary ACPs to synthesize second-stranded cDNAs under conditions such that the 3′-end core portion of dT-ACP2 is prevented from annealing to single-stranded cDNAs and only the 3′-end core portion of the arbitrary ACP anneals to the single-stranded cDNAs. Arbitrary ACPs contain random 10-mers as the 3′-end core sequences. Only those ACPs that are sufficiently complementary to a region of a first-strand cDNA were expected to anneal and be amplified. Two microliters of the PCR products ware separated by electrophoresis on 2% agarose gels. Bands showing different amounts of the resultant PCR products were extracted by the DE81 paper (QIAGEN) method, cloned into a pGEM-T easy vector (Promega, Madison, WI), and then sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and ABI 7700 DNA sequencer. After identification of clones by BLAST analysis, Semiquantitative PCR using gene-specific primers (Table 1) and the pooled samples was carried out to select candidate genes that were differentially expressed between the narcolepsy and the healthy control patients. The band intensities were determined using the NIH Image 1.61 software (National Institute of Health, Bethesda, MD). Human beta 2-microglobulin (B2M) was used as a control to confirm the integrity of the total RNA samples. Genes that showed a difference of more than 2-fold in their expression intensity were considered to be differentially expressed. The identified candidate genes were then further studied by quantitative real-time PCR by using a new set of cases without regarding disease duration (20 narcolepsy-cataplexy patients and 20 age- and sex-matched healthy controls). The expression levels of these identified genes were determined with the ABI 7300 RT-PCR system using SYBR Green (TAKARA, Japan) and gene-specific primers. Single-stranded cDNA was synthesized using the Superscript III first-strand synthesis system with random hexamer (Invitrogen, Carlsbad, CA). According to the Applied Biosystems guide to performing relative quantitation of gene expression using real-time quantitative PCR, the relative quantity of target gene expression was evaluated using the comparative threshold method with beta 2-microglobulin as an internal control. Distributions of relative gene expressions were compared between the narcolepsy group (n = 20) and the healthy control group (n = 20) using the Mann-Whitney U test. A value of P <0.05 was considered to be significant.
Table 1.
Primer Sequences Used in Semiquantitative RT-PCR and Real-Time RT-PCR.
| Gene Symbol | Accession Number | Gene-Specific Primers |
|---|---|---|
| MX2 (MxB) | M30818 | 5′-TCTGTCACTATCAGTGTCCATCTCTAC (*2) |
| 5′-TCTTTGCTTTATTAAATTCCTCTTCAA (*1) | ||
| 5′-AGAAATTACATTCTTTCAAACACATCC (*2) | ||
| 5′-GATCTCAAATGTCTTGTAGTTGACAAA (*2) | ||
| ATP6V1E1 | BC004443 | 5′-AGAAGAAGAGTTCAACATAGAGAAAGGT |
| 5′-ATTTAGTAGGTCTGTGATAAGGTCATCTCT | ||
| GMFG | BC101818 | 5′-CTAGAAGACAGCGGAACTAAGAAAAG |
| 5′-GTTTTTACTCCCTGCATACATCATC | ||
| IFI6 | BC015603 | 5′-GTTCTCACTATATTGTCCAGGCTAGAGT |
| 5′-AGTTTATTCTGTTTTCACATCTAGGTTGTT | ||
| PRPF8 | BC064370 | 5′-GAATCTATGAAGTGGAAGAAGCTAACTAAT |
| 5′-ATTGTCTCCTTTTGTACTGTCTCAATTT | ||
| c19orf43 | BC000216 | 5′-AGGATGAGGTATTAACAAGTAAAGGTGAC |
| 5′-GACAGTAAACCACTGTTCTATGAAGTCTC | ||
| c20orf77 | BC033629 | 5′-ATTAATGAGTCTATAAGGTTTTTCTTCCAG |
| 5′-ATTCATTTACATAATTGCTACAGTTTCATT | ||
| NALP1 | BC051787 | 5′-GTCAGGAATTTACTACACAGAAATCAGA |
| 5′-CAAATAAGTCTCTGATCTCAATTAAATGTC | ||
| PARP6 | BC110902 | 5′-AAGATTCTTCTTTGTATATGAGGATGGT |
| 5′-ATTTTATTGTTTTATTTACAAACAGGGTGA | ||
| FLJ45432 | AK127358 | 5′-GATGGTGATTGATCCTTTGACAC |
| 5′-GCTGGAAAAAGGAAGCACTCT | ||
| B2M | BC032589 | 5′-TGCTGTCTCCATGTTTGATGTATCT |
| 5′-TCTCTGCTCCCCACCTCTAAGT |
(*1) 3′ specific region primer for MxB; (*2) common region primers for MX2 and MxB
The time of blood collection was generally between 14:00 and 16:00, although some samples were collected in the morning or in the evening. To examine the circadian variations in the expression levels of the candidate genes, we developed a distribution graph and conducted an analysis of variance (ANOVA) test by dividing all subjects (both narcolepsy and control patients) into 3 groups, depending on the time of blood sampling, and checked whether there were any differences in the relative gene expression level between these groups. Blood was drawn between 08:00 and 12:00 in the first group, between 12:01and 16:00 in the second group, and between 16:01 and 20:00 in the third group.
RESULTS
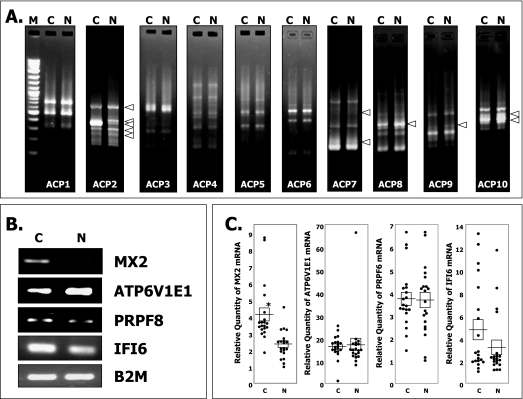
By using the first 10 most efficient commercially available 10 primers (ACP1 to 10), we successfully cloned 11 PCR products that appeared to be differentially expressed between the pooled RNA samples from the narcolepsy patients and those from the healthy controls (Figure 1A). Eleven clones were sequenced and identified as follows: [myxovirus (influenza virus) resistance 2] (MX2) (Acc#: BC035293), [ATPase, H+ transporting, lysosomal 31 kDa, V1 subunit E1] (ATP6V1E1) (Acc#: BC004443), [glia maturation factor gamma] (GMFG) (Acc#: BC101818), [interfer on (IFN)-inducible peptide precursor] (IFI6) (Acc#: BC015603), [pre-mRNA processing factor 8] (PRPF8) (Acc#: BC064370), [c19orf43] (Acc#: BC000216), [c20orf77] (Acc#: BC033629), [NACHT, leucine rich repeat and PYD (pyrin domain) containing 1] (NALP1) (Acc#: BC051787), [poly(ADP-ribose) polymerase family, member 6] (PARP6) (Acc#: BC110902), [FLJ45432] (Acc#: AK127358), and [IFN-induced cellular resistance mediator protein] (MxB) (Acc#: M30818). Among them, 2 selected genes, i.e., MxB and MX2, happened to be a splice variant of the same gene. To verify the expressional difference of the selected genes of interest, semiquantitative RT-PCR was carried out with the same pooled sample using gene-specific primers (Table 1). Four candidate genes—MX2, ATP6V1E1, IFI6, and PRPF8— were differentially expressed between the 2 groups (Figure 1B).
Figure 1.
Differentially expressed genes in narcoleptic white blood cells. (A) Representative PCR band patterns for genes that are differentially expressed between healthy subjects and patients with narcolepsy using annealing control primers and pooled-RNA samples. Equal amounts (3 μg) of pooled total RNA was used as a reverse transcription-polymerase chain reaction (RT-PCR) template. The PCR products were separated by electrophoresis on 2% agarose gels. M, lane containing a 100-bp DNA ladder marker; C, healthy control subject; N, patients with narcolepsy. (B) Semiquantitative PCR products obtained with 30 cycles using gene-specific primers and pooled total RNA samples for further confirmation. MX2, myxovirus resistance 2; ATP6V1E1, ATPase, H+ transporting, lysosomal 31 kDa, V1 subunit E1; IFI6, interferon-inducible peptide precursor; PRPF8, premRNA processing factor 8; B2M, beta-2 microgloblin. (C) Quantitative real-time PCR analysis with gene-specific primers using the delta-delta cycle threshold method. Each dot corresponds to the relative value of the gene expression level in each subject. Horizontal lines indicate the mean of each group and squares indicate the standard error of each group. The asterisk shows the significant difference between the 2 groups (P <0.0005)
With regard to MX2, we conducted PCR amplification using 2 sets of primers to determine if the differentially expressed clones were MxB, MX2, or both. One pair of primers (the top 2 lines in Table 1) includes the MxB-specific 3′ region and the other pair (the next 2 lines in Table 1) is located in the common region of MX2 and MxB from nucleotide positions 2522 to 2679 on MxB. We found that both clones (MxB and MX2) were amplified, and the relative expression levels of the MX2 and MxB clones in 40 individuals showed a strong correlation (Spearman's correlation coefficient test: n = 40, R2 = 0.7848), suggesting that 2 PCR products were amplified from a single gene. There were absolutely no differences in the registered open reading frame sequences of MX2 and MxB. MX2, which is registered as a representative clone in GenBank, might just be a shorter isoform of MxB, not a full-length clone. Therefore, we used 1 primer set from the common region of MX2 and MxB for the further analysis of these genes.
The 4 candidate genes that were found to be differentially expressed were further studied by quantitative real-time PCR analysis. Among them, 1 gene, i.e., MX2, was clearly down-regulated in patients with narcolepsy, and the U test showed a significant difference between the narcolepsy (mean ±̣ SD; 2.32 ±̣ 0.83) and control groups (mean ±̣ SD; 4.10 ±̣ 1.77) (Figure 1C,P <0.00005). The relative MX2 expression levels in 3 healthy controls with HLA-DRB1*1501-DQB1*0602 were 3.45, 3.66, and 3.82 and were within the mean ± twice the standard error (4.10 ±̣ 0.79) values of the healthy subjects. In order to evaluate the effect of circadian variation on MX2 expression, we conducted an ANOVA test between the 3 groups of subjects that were divided according to the blood sampling time and found no significant difference between the groups (P = 0.14).
DISCUSSION
In this study, we applied the new ACP technology to RNA extracted from blood cells and successfully identified the MX2 gene as a dysregulated gene in narcoleptic patients. This is the first report to identify the biological markers of narcolepsy using gene expression in white blood cells. Recent advances in narcolepsy research have revealed the importance of the hypocretin system, but it remains to be clarified why narcolepsy shows a very close HLA association and whether an immunological alteration underlies the pathophysiology of narcolepsy. Our method that involves the use of mRNA from white blood cells has the advantage of detecting immune-related narcolepsy genes. Although gene expression in white blood cells is expected to vary greatly according to an individual's health condition, using a pooled-sample strategy can reduce individual variations and enhance group-specific expression changes. Therefore, genes that are consistently upregulated or downregulated in the groups can be efficiently detected.
The human genome contains 2 conserved IFN-regulated genes—MX1 and MX2. These gene are specifically induced by IFN-alpha and beta.10,11 MX proteins belong to the family of large GTPases and are highly homologous with dynamins within their GTP-binding domain. MX proteins have a conserved C-terminal leucine zipper element. MX2 exists in 78-kDa and a 76-kDa forms, and these 2 molecules form hetero-oligomers via the C-terminal leucine zipper.12 They are translocated into the nucleus under IFN-alpha stimulation. Although MX2 exists both in the nucleus and cytoplasm, no colocalization/oligomerization with the cytoplasmic MX1 was demonstrated. Each protein might have differential antiviral activities in the nucleus or cytoplasm, whereas no antiviral activity has been demonstrated by the human MX2 protein yet. In the absence of IFNs, both MX1 and MX2 form inactive oligomeric molecules. On viral challenge (such as bunyavirus infection), antivirally competent MX1 monomers are released from these oligomeric molecules and bind to viral targets.13 Although no specific viral targets of MX2 have been identified, it is conceivable that MX2 monomers also exhibit antiviral properties by binding to their own viral targets, which have not yet been identified.12
It is also known that MX genes are highly polymorphic among species, indicating differential antiviral activities.14,15 In mice, all feral strains examined show the expression of MX2 mRNA, but no laboratory mice have detectable levels of the MX2 gene15 because the single nucleotide adenine insertion in laboratory mice results in premature termination of MX2 protein synthesis. Unexpectedly, we found that this undetectable level of MX2 mRNA in laboratory mice was probably caused by nonsense mRNA-mediated decay.16,17 Human MX2 might also be polymorphic, exhibiting different antiviral activity according to racial and/or residential differences.
Two IFN-inducible MX proteins, MX1 and MX2, are located on human chromosome 21q22.3. Recently 2 genome-wide association studies indicated a strong association between human narcolepsy and chromosome 21q22.3.18,19 Kawashima et al have already identified candidate genes in the 21q22.3 gene locus.19 The nearest marker rs12483718 is different from the MXs loci. No candidate genes have been identified from the narcolepsy susceptibility gene locus reported by Dauvillier et al. The locus is located on 21q22.3 from the ABCG1 gene to the D21S267 genetic marker,18 and this 5.2 Mb region contains 2 MX loci. Therefore, MX2 might be a novel susceptibility gene for narcolepsy as suggested by a human narcolepsy study in a large French family.18
This study identifies a novel gene, MX2, which shows clear transcriptional changes in narcolepsy; however, the direct relevance of this gene in narcolepsy has not been elucidated. Further study is needed to explore the functional relationship between the MX2 gene and narcolepsy and characterize the effect of IFNs in narcolepsy. Type I IFNs (IFN-alpha and beta) are potent antiviral cytokines and modulators of the adaptive immune system. It should be noted that the activity of the prepro-orexin promoter is modulated by IFN-alpha.20 It may be possible that narcoleptic patients have different IFN status or responsiveness against IFNs as compared with healthy subjects. In this context, a technical improvement for IFN measurement is required because conventional enzyme-linked immunosorbent assay detects not only biologically active forms but also inactive form of IFNs.
The other candidate genes selected are also of potential interest because they may indicate the factors related to narcolepsy. However, like MX2, their functions are sometimes hypothetical and have not always been elucidated. Among these genes, some are implicated in viral infection. IFI6 is transcriptionally induced by IFN-alpha and beta,11,21,22 virus infections,23–25 and TNF26, however, the precise functions of IFI6 remain unknown. ATPV1E1 is one component of the ATP catalytic site of the vacuolar proton pump (V-ATPase). V-ATPase is responsible for the acidification of endosomes and transportation of hydrogen.27 Endosome acidification has been observed in virus infections such as influenza.28 Acidification of endosomes stimulates fusion of the viral and endosomal membranes; this causes a structural change in the viral homotrimeric glycoprotein hemagglutinin. An abnormality in one subunit on V-ATPase might be relevant to viral infection. PRPF8 is a component of mammalian spliceosomes, which are large multiprotein complexes involved in the removal of introns from mRNA precursors.29 Mutations in PRPF8 have been observed in autosomal dominant retinitis pigmentosa.30 The relationship between narcolepsy and PRPF8 has not been addressed yet.
A limitation of this study is that the HLA-DRB 1*1501-DQB1*0602 haplotype was detected only in approximately 20% of our control subjects. The use of HLA-matched controls would have yielded better results. We believe that the change in the MX2 expression levels observed here is not solely due to the difference in HLA haplotypes, because these levels in the HLA-DRB1*1501-DQB1*0602-positive controls were close to the mean of the healthy control subjects (Figure 1C). Another limitation is that we did not collect the cerebral spinal fluid (CSF) from the subjects for analysis. CSF hypocretin measurement would be necessary to clarify the relationship between MX2 expression and hypocretin status.
In conclusion, we show here, for the first time, a downregulation of MX2 in narcoleptic white blood cells by using a new ACP technology. MX2 is known to be influenced by IFN status and has potential antiviral activity. Further studies are required to determine its functional relationship to the pathophysiology of narcolepsy. Analysis using this new technology reveals the gene expression changes specific for narcolepsy. Applying this technology to other hypersomnias might be helpful in identifying biological markers useful for the differential diagnosis of narcolepsy in the future.
ACKNOWLEDGMENTS
We thank Ms. Junko Watanabe (Neuropsychiatric Research Institute, Japan) for clinical coordination and Ms. Miyuki Fukazawa (Tokyo Institute of Psychiatry) and Ms. Yoshino Kanai (Neuropsychiatric Research Institute, Japan) for blood processing. This work was supported by Grants-in-Aid for Scientific Research (No. 16659308, No. 17390324, and No. 17700362) and on Priority Areas “Comprehensive Genomics” from the Ministry of Education, Science and Culture of Japan.
Footnotes
Disclosure Statement
This was not an industry supported study. Drs. Tanaka, Yutaka Honda, and Makoto Honda have indicated no financial conflicts of interest.
REFERENCES
- 1.American Academy of Sleep Medicine. diagnostic & coding manual. 2nd ed. Westchester, IL: American Academy of Sleep Medicine; 2005. International classification of sleep disorders. [Google Scholar]
- 2.Scammell TE. The frustrating and mostly fruitless search for an autoimmune cause of narcolepsy. Sleep. 2006;29:601–2. doi: 10.1093/sleep/29.5.601. [DOI] [PubMed] [Google Scholar]
- 3.Vgontzas AN, Bixler EO, Lin HM, Prolo P, Trakada G, Chrousos GP. IL-6 and its circadian secretion in humans. Neuroimmunomodulation. 2005;12:131–40. doi: 10.1159/000084844. [DOI] [PubMed] [Google Scholar]
- 4.Okun ML, Giese S, Lin L, Einen M, Mignot E, Coussons-Read ME. Exploring the cytokine and endocrine involvement in narcolepsy. Brain Behav Immun. 2004;18:326–32. doi: 10.1016/j.bbi.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Hohjoh H, Nakayama T, Ohashi J, et al. Significant association of a single nucleotide polymorphism in the tumor necrosis factor-alpha (TNF-alpha) gene promoter with human narcolepsy. Tissue Antigens. 1999;54:138–45. doi: 10.1034/j.1399-0039.1999.540204.x. [DOI] [PubMed] [Google Scholar]
- 6.Hohjoh H, Terada N, Kawashima M, Honda Y, Tokunaga K. Significant association of the tumor necrosis factor receptor 2 (TNFR2) gene with human narcolepsy. Tissue Antigens. 2000;56:446–8. doi: 10.1034/j.1399-0039.2000.560508.x. [DOI] [PubMed] [Google Scholar]
- 7.Himmerich H, Beitinger PA, Fulda S, et al. Plasma levels of tumor necrosis factor alpha and soluble tumor necrosis factor receptors in patients with narcolepsy. Arch Intern Med. 2006;166:1739–43. doi: 10.1001/archinte.166.16.1739. [DOI] [PubMed] [Google Scholar]
- 8.Dauvilliers Y, Carlander B, Rivier F, Touchon J, Tafti M. Successful management of cataplexy with intravenous immunoglobulins at narcolepsy onset. Ann Neurol. 2004;56:905–8. doi: 10.1002/ana.20339. [DOI] [PubMed] [Google Scholar]
- 9.Arnulf I, Lin L, Zhang J, et al. CSF versus serum leptin in narcolepsy: is there an effect of hypocretin deficiency? Sleep. 2006;29:1017–24. doi: 10.1093/sleep/29.8.1017. [DOI] [PubMed] [Google Scholar]
- 10.Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 11.Kelly JM, Porter AC, Chernajovsky Y, Gilbert CS, Stark GR, Kerr IM. Characterization of a human gene inducible by alphaand beta-interferons and its expression in mouse cells. Embo J. 1986;5:1601–6. doi: 10.1002/j.1460-2075.1986.tb04402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melen K, Julkunen I. Nuclear cotransport mechanism of cytoplasmic human MxB protein. J Biol Chem. 1997;272:32353–9. doi: 10.1074/jbc.272.51.32353. [DOI] [PubMed] [Google Scholar]
- 13.Haller O, Kochs G. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–7. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- 14.Ko JH, Jin HK, Asano A, et al. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 2002;12:595–601. doi: 10.1101/gr.210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin HK, Takada A, Kon Y, Haller O, Watanabe T. Identification of the murine Mx2 gene: interferon-induced expression of the Mx2 protein from the feral mouse gene confers resistance to vesicular stomatitis virus. J Virol. 1999;73:4925–30. doi: 10.1128/jvi.73.6.4925-4930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsensemediated decay approaches the clinic. Nat Genet. 2004;36:801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 17.Inoue K, Khajavi M, Ohyama T, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–9. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 18.Dauvilliers Y, Blouin JL, Neidhart E, et al. A narcolepsy susceptibility locus maps to a 5 Mb region of chromosome 21q. Ann Neurol. 2004;56:382–8. doi: 10.1002/ana.20208. [DOI] [PubMed] [Google Scholar]
- 19.Kawashima M, Tamiya G, Oka A, et al. Genomewide association analysis of human narcolepsy and a new resistance gene. Am J Hum Genet. 2006;79:252–63. doi: 10.1086/505539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waleh NS, Apte-Deshpande A, Terao A, Ding J, Kilduff TS. Modulation of the promoter region of prepro-hypocretin by alpha-interferon. Gene. 2001;262:123–8. doi: 10.1016/s0378-1119(00)00544-8. [DOI] [PubMed] [Google Scholar]
- 21.Hibbert L, Foster GR. Human type I interferons differ greatly in their effects on the proliferation of primary B cells. J Interferon Cytokine Res. 1999;19:309–18. doi: 10.1089/107999099314009. [DOI] [PubMed] [Google Scholar]
- 22.Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984;38:745–55. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 23.Clauss IM, Wathelet MG, Szpirer J, et al. Chromosomal localization of two human genes inducible by interferons, double-stranded RNA, and viruses. Cytogenet Cell Genet. 1990;53:166–8. doi: 10.1159/000132920. [DOI] [PubMed] [Google Scholar]
- 24.Wathelet MG, Clauss IM, Paillard FC, Huez GA. 2-Aminopurine selectively blocks the transcriptional activation of cellular genes by virus, double-stranded RNA and interferons in human cells. Eur J Biochem. 1989;184:503–9. doi: 10.1111/j.1432-1033.1989.tb15043.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Zhao H, Collins CD, et al. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37:1180–8. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]
- 26.Chernajovsky Y, Reid TR. Regulation of the human interferon-inducible 6-16 promoter in tumor necrosis factor-sensitive and resistant mouse cells: role of cAMP as a mediator of signal transduction. J Interferon Res. 1990;10:627–36. doi: 10.1089/jir.1990.10.627. [DOI] [PubMed] [Google Scholar]
- 27.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–89. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 28.Huang Q, Sivaramakrishna RP, Ludwig K, Korte T, Bottcher C, Herrmann A. Early steps of the conformational change of influenza virus hemagglutinin to a fusion active state: stability and energetics of the hemagglutinin. Biochim Biophys Acta. 2003;1614:3–13. doi: 10.1016/s0005-2736(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 29.Luo HR, Moreau GA, Levin N, Moore MJ. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. Rna. 1999;5:893–908. doi: 10.1017/s1355838299990520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10:1555–62. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]