Abstract
Marked sarcomere disorganization is a well-documented characteristic of cardiomyocytes in the failing human myocardium. Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC2v), which is involved in the development of human cardiomyopathy, is an important structural protein that affects physiologic cardiac sarcomere formation and heart development. Integrated cDNA expression analysis of failing human myocardia uncovered a novel protein kinase, cardiac-specific myosin light chain kinase (cardiac-MLCK), which acts on MLC2v. Expression levels of cardiac-MLCK were well correlated with the pulmonary arterial pressure of patients with heart failure. In cultured cardiomyocytes, knockdown of cardiac-MLCK by specific siRNAs decreased MLC2v phosphorylation and impaired epinephrine-induced activation of sarcomere reassembly. To further clarify the physiologic roles of cardiac-MLCK in vivo, we cloned the zebrafish ortholog z–cardiac-MLCK. Knockdown of z–cardiac-MLCK expression using morpholino antisense oligonucleotides resulted in dilated cardiac ventricles and immature sarcomere structures. These results suggest a significant role for cardiac-MLCK in cardiogenesis.
Introduction
Despite recent advances in pharmacologic and surgical therapies, chronic heart failure (CHF) is still a leading cause of death worldwide (1). Currently, heart transplant is thought to be the most effective therapy for end-stage CHF. However, this approach obviously cannot be used for all of the numerous affected patients and is not suitable for patients with a mild disease state. Therefore, there is increasing demand for new therapeutic targets for CHF.
Cardiomyocytes, the most basic cellular unit of the myocardium, express several sarcomeric proteins, including myosin and actin; abnormalities in these sarcomeric proteins are major causes of idiopathic cardiomyopathies and lead to CHF (2–4). Type II myosin is the major constituent of sarcomeres. In the neck region of this protein, there are binding sites for a pair of myosin light chains, which are called the essential light chain and the regulatory light chain. Among the several paralogs of the myosin regulatory light chain in vertebrates (5), myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC2v) is expressed in the myocardium, where it performs specific roles in cardiogenesis by contributing to the formation of sarcomeres and in increasing the Ca2+ sensitivity of muscle tension at submaximal Ca2+ concentrations (6, 7). Currently, 2 members of the myosin light chain kinase (MLCK) protein family that act on myosin regulatory light chain in muscle cells have been identified, skeletal muscle MLCK (skMLCK) and smooth muscle MLCK (smMLCK) (8). Among these MLCK family members, smMLCK, including nonmuscle isoforms, is distributed ubiquitously in various tissues and contributes to the contraction of smooth muscle and several cell activities. Conversely, skMLCK is thought to localize and function in both cardiac muscle and skeletal muscle (9); to our knowledge, no cardiac-specific MLCK has been reported to date. skMLCK-deficient mice, however, did not show any heart weight, body weight, or heart weight/body weight ratio phenotypes, despite effective knockdown of skMLCK expression (10). Additionally, there were no significant differences between the knockout and wild-type animals in regard to MLC2v phosphorylation, suggesting the existence of as-yet unknown kinases in cardiac muscle cells.
Genome-wide analyses, which have recently become available in a wide range of clinical settings, such as cancer research, allow for a global view of gene expression in certain disease states and the identification of unknown molecules and molecular pathways that can be exploited as novel therapeutic targets. CHF is a candidate disease for this type of genome-wide analysis, because of its heterogeneous properties and previous difficulties identifying responsible genes using other conventional modalities.
In this study, we performed microarray analysis of the failing human myocardium and examined the correlation between the obtained genomic data and the clinical, physiological, and biochemical characteristics of CHF. In this manner, we sought to identify candidate genes that are involved in the pathophysiology of CHF. Consequently, we identified what we believe to be a novel cardiac-specific MLCK (cardiac-MLCK; encoded by MYLK3). Phosphorylation of MLC2v by cardiac-MLCK regulated the reassembly of sarcomere structures in cultured neonatal rat cardiomyocytes. Suppression of cardiac-MLCK expression in zebrafish embryos using specific morpholino antisense oligonucleotides (MOs) led to dilation of the cardiac ventricle with incomplete sarcomere formation, suggesting critical roles for cardiac-MLCK in the heart.
Results
Identification of cardiac-MLCK from failing human myocardia using microarray analysis.
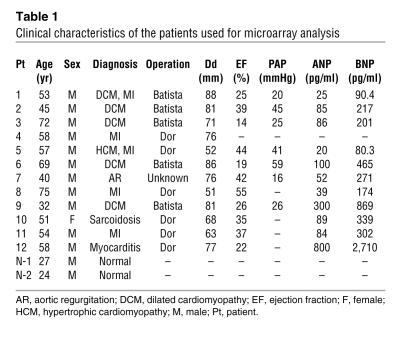
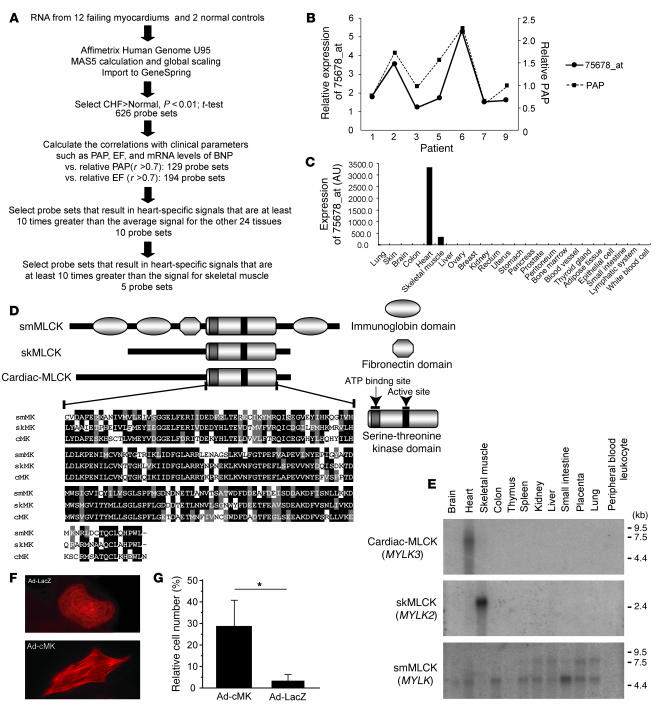
To identify candidate genes involved in the pathophysiology of CHF, we used an HG-U95 Affymetrix GeneChip to analyze the gene expression profiles of failing myocardial tissues obtained from 12 patients who had undergone cardiac exclusion surgery, such as the Dor or Batista procedures, for end-stage CHF (Table 1). Figure 1A is an overview flowchart for the selection of candidate genes. Compared with those of 2 normal control samples, the expression of 626 probe sets was significantly upregulated in the failing myocardia. Of these, we selected probe sets whose expression levels were positively correlated (r > 0.7) with pulmonary arterial pressure (PAP) measurements (129 probe sets) and brain natriuretic peptide (BNP) mRNA levels (194 probe sets). The tissue localization of each selected probe set was then analyzed using the commercially available BioExpress database (Gene Logic Inc.). We selected 10 probe sets, for which the cardiac expression level was at least 10-fold the mean expression level of 24 other tissues, for further analysis. These probe sets represented a set of genes that included atrial natriuretic peptide (ANP), BNP, small muscle protein, and α-actin, all of which are known to be involved in heart failure, cardiac muscle remodeling, and striated muscle function. We calculated the ratios of expression in cardiac muscle to that in skeletal muscle in these probe sets. ANP (36663_at and 73106_s_at), BNP (39215_at), Importin9 (84730_at), and 75678_at exhibited expression levels that were at least 10-fold greater in the heart than in skeletal muscle. Expression levels of 75678_at, for which annotation was not available, were similar to those of ANP and BNP. We hypothesized that this unknown transcript was involved in the pathophysiology of heart failure.
Table 1 .
Clinical characteristics of the patients used for microarray analysis
Figure 1. Microarray analysis for candidate gene selection.
(A) Flowchart for the selection of candidate genes. (B) The relative expression levels of 75678_at correlated well with the relative PAP values in the respective patients. (C) Tissue localization of the candidate gene expression was analyzed using the GeneExpress database; 75678_at was specifically expressed in the heart. (D) Each MLCK family member possesses a highly conserved serine-threonine kinase domain in the C-terminal region of the protein. Amino acid residues on black backgrounds are the most commonly conserved residues at each position; residues on gray backgrounds are similar to the consensus amino acids. (E) Expression analysis of MLCK family members using multiple human tissue Northern blot membranes. The 2 transcripts transcribed from MYLK (encoding smMLCK) were ubiquitously expressed with the exception of skeletal muscle, thymus, and peripheral blood leukocytes. In contrast, MYLK2 (encoding skMLCK) and MYLK3 (encoding cardiac-MLCK) were only expressed in skeletal muscle and heart, respectively. (F) Fluorescence microscopy of cardiomyocytes cultured in serum-free conditions and infected with adenovirus encoding LacZ (Ad-LacZ) revealed predominantly round-shaped cells with disorganized sarcomere structures. Infection with adenovirus encoding cardiac-MLCK (Ad-cMK) at a MOI of 120 increased the number of the cells with organized sarcomere structures. Original magnification, ×1,000. (G) The percentage of cells with organized sarcomeres was significantly higher in cardiomyocytes infected with adenovirus encoding cardiac-MLCK than in those infected with adenovirus encoding LacZ. Values are mean ± SEM. *P < 0.001.
Using 5′-RACE, we identified specific sequences identical to those of NM_182493 (MYLK3) located 4 kb upstream of the probe set sequence. The relative expression level of this candidate gene was significantly correlated with the relative PAP value (Figure 1B); in addition, the expression of this gene was restricted to the heart (Figure 1C). A homology search using the transcript sequence, particularly the sequence coding for the C-terminal kinase domain, identified MYLK3 as a member of the MLCK family. Thus, we named the protein encoded by MYLK3 “cardiac-MLCK.” Two distinct MLCK family genes have been previously reported: MYLK, which encodes smMLCK, and MYLK2, which encodes skMLCK (8). Domain structure analysis revealed a well-conserved serine/threonine kinase domain that includes an ATP-binding site and an active serine/threonine kinase domain positioned near the C terminus of the cardiac-MLCK protein (Figure 1D). The expression patterns of the MLCK family members were confirmed by Northern blot analysis. As previously described (11), 2 major transcripts of MYLK were almost ubiquitously expressed. The larger transcript codes for a nonmuscle isoform of smMLCK generated by alternative splicing. Restricted expression patterns were observed for both MYLK2 and MYLK3. MYLK2 expression was only detected in skeletal muscle, whereas MYLK3 expression was only observed in the heart (Figure 1E). MYLK was also found to be expressed in the heart, although its expression was not upregulated in failing myocardia as much as the expression of MYLK3 (data not shown). To assess the physiological significance of cardiac-MLCK, we generated an adenovirus vector encoding cardiac-MLCK. In serum-free conditions, cultured neonatal rat cardiomyocytes showed predominantly disorganized sarcomere structures. Overexpression of cardiac-MLCK in cultured neonatal rat cardiomyocytes augmented sarcomere organization under serum-starved conditions (cells with organized sarcomeres, 28.7% ± 11.1% versus 3.1% ± 2.4%; P < 0.001; Figure 1, F and G), suggesting that cardiac-MLCK participates in sarcomere formation in cardiomyocytes.
Cardiac-specific myosin regulatory light chain is a specific substrate of cardiac-MLCK.
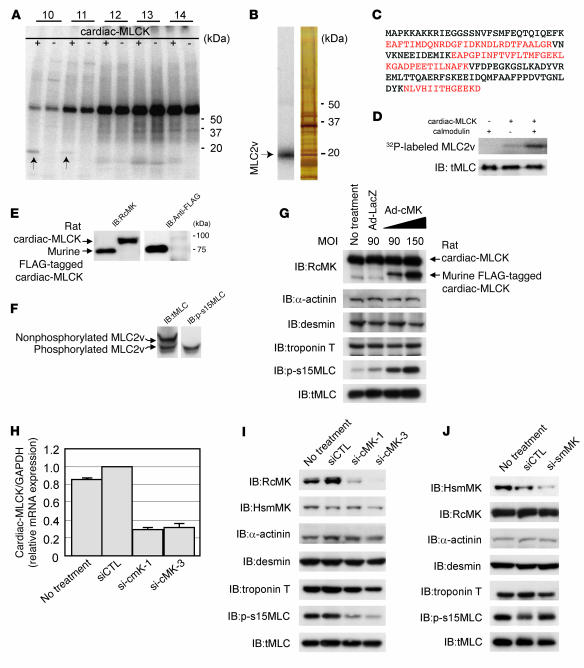
Because this protein kinase contained a consensus kinase catalytic domain, we attempted to identify potential substrates of cardiac-MLCK. To identify physiological substrates of cardiac-MLCK, we screened murine heart homogenates using an in vitro kinase reaction. After fractionation of murine heart homogenates using a cation exchange column, aliquots of each fraction were subjected to an in vitro kinase reaction with recombinant cardiac-MLCK. Fractions 10 and 11 each contained a distinct 20-kDa band that was labeled with 32P only in the presence of recombinant cardiac-MLCK (Figure 2A). This 32P-labeled 20-kDa protein was purified (Figure 2B) and analyzed using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry and peptide mass fingerprinting. The 20-kDa protein contained fragments with amino acid sequences that were homologous to murine MLC2v (Figure 2C). No additional 32P-labeled proteins were detected in fractions obtained following cation or anion exchange column purification. Further analysis of this phosphorylation event in vitro revealed endogenous MLC2v, purified from murine heart homogenates, was phosphorylated by recombinant cardiac-MLCK in a Ca2+-calmodulin–dependent manner (Figure 2D). Thus, we conclude that cardiac-MLCK is a calmodulin-dependent kinase.
Figure 2. Identification of MLC2v as a specific substrate of cardiac-MLCK.
(A) A putative 20-kDa substrate that was labeled with P32 in the presence of cardiac-MLCK was identified in fractionated murine myocardium extracts (arrows). Fraction numbers are shown at top. (B) P32-labeled MLC2v was purified and visualized by autoradiography (left lane) and silver staining (right lane). (C) Peptides from the purified protein, which matched the sequences of murine MLC2v, are shown in red. (D) Purified MLC2v from murine myocardia was phosphorylated by cardiac-MLCK in a Ca2+-calmodulin–dependent manner. (E) RcMK detected rat cardiac-MLCK from cultured cardiomyocyte cell extracts and FLAG-tagged murine cardiac-MLCK. (F) Nonphosphorylated MLC2v and phosphorylated MLC2v were separated using urea-glycerol gel electrophoresis. tMLC and p-s15MLC were confirmed to specifically detect each target protein. (G) Overexpression of murine cardiac-MLCK in cultured cardiomyocytes following infection with an adenovirus vector encoding murine cardiac-MLCK at MOIs of 90 and 150 upregulated the phosphorylation of MLC2v in a dose-dependent manner. Endogenous rat cardiac-MLCK is shown at top; overexpressed murine cardiac-MLCK is shown below. (H and I) Both si-cMK-1 and si-cMK-3 effectively suppressed the mRNA (H) and protein levels (I) of cardiac-MLCK, resulting in reduced phosphorylation of MLC2v. smMLCK, α-actinin, desmin, and troponin T were not affected by suppression of cardiac-MLCK expression. siCTL, control siRNA. (J) The protein levels of smMLCK were effectively decreased by si-smMK; no remarkable changes were observed in protein levels of phosphorylated MLC2v or other sarcomere-related proteins.
Next, we generated polyclonal antibodies specific for rodent cardiac-MLCK (RcMK). Antibodies that detected phosphorylated MLC2v (p-s15MLC; anti-rodent serine 15 phosphorylated MLC2v) and total MLC2v (tMLC) were also generated. RcMK detected rat cardiac-MLCK from whole-cell cardiomyocyte extracts as well as recombinant FLAG-tagged murine cardiac-MLCK (Figure 2E). Phosphorylated MLC2v and nonphosphorylated MLC2v could be clearly separated using urea-glycerol gel electrophoresis (12). tMLC detected both phosphorylated and nonphosphorylated MLC2v, whereas p-s15MLC specifically detected the phosphorylated form of MLC2v (Figure 2F). Overexpression of cardiac-MLCK increased the levels of phosphorylated MLC2v in cultured cardiomyocytes (Figure 2G). However, there was no effect on the expression of other sarcomere proteins involved in sarcomere organization such as troponin T, desmin, and α-actinin. mRNA expression of ANP and β myosin heavy chain, representative markers of cardiac hypertrophy, were also unaffected by cardiac-MLCK overexpression (data not shown). To further investigate the phosphorylation of MLC2v by endogenous cardiac-MLCK, we used specific siRNAs targeting cardiac-MLCK (si-cMKs). These siRNAs effectively suppressed the level of cardiac-MLCK mRNA by more than 70%, as determined using quantitative real-time PCR 24 hours after transfection (Figure 2H). These siRNAs also effectively suppressed the level of cardiac-MLCK protein and the amount of phosphorylated MLC2v 60–72 hours after transfection (Figure 2I), whereas no remarkable effects were seen for the expression of other sarcomere proteins. On the contrary, suppression of smMLCK expression, which is also distributed in heart, using siRNA targeting rat smMLCK (si-smMK) did not change either the phosphorylation status of MLC2v or the expression of sarcomere proteins (Figure 2J). These results indicated that cardiac-MLCK predominantly phosphorylates MLC2v, which is selectively expressed in cardiomyocytes. Thus, cardiac-MLCK may regulate morphologic change in cardiomyocytes, including sarcomere organization, through MLC2v phosphorylation.
Cardiac-MLCK regulates sarcomere assembly in cultured cardiomyocytes.
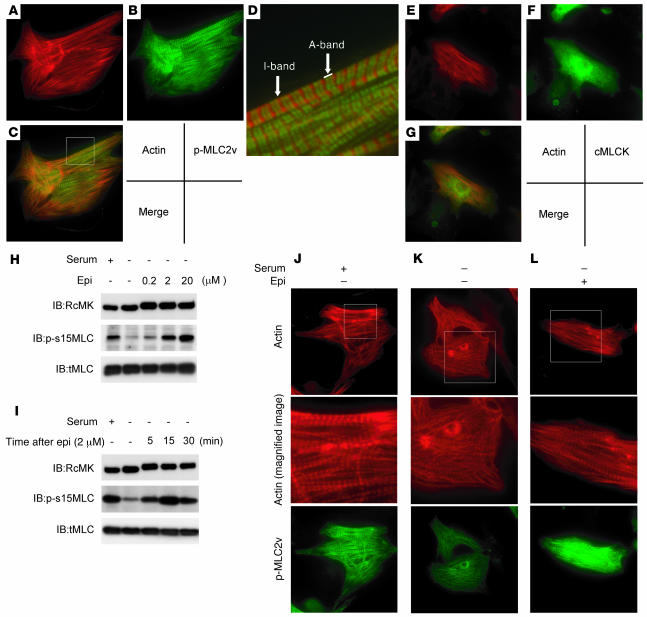
To elucidate the precise role of cardiac-MLCK in the sarcomere structure, we analyzed the effects of MLC2v phosphorylation on sarcomeres in cultured neonatal rat cardiomyocytes. Polymerized actin stained with rhodamine-phalloidin revealed a regularly organized pattern of striations (Figure 3A). Phosphorylated MLC2v labeling with p-s15MLC demonstrated a similar striated pattern, although the labeling was predominantly observed in the A-band region, a portion of the sarcomere primarily made up of thick filaments (Figure 3, B–D). Diffuse cytosolic fluorescent labeling was seen when cardiac-MLCK was labeled with RcMK (Figure 3, E–G).
Figure 3. Epinephrine treatment induced sarcomere assembly through MLC2v phosphorylation.
Original magnification, ×1,000 (A–C and E–G). (A–D) Polymerized actin stained with rhodamine-phalloidin (A) as well as phosphorylated MLC2v labeled with p-s15MLC (B) exhibited regular patterns of striation. (C) Merged image of A and B. (D) Higher magnification of boxed area in C revealed that rhodamine-phalloidin predominantly stained the I-band, whereas phosphorylated MLC2v (p-MLC2v) was localized in the A-band. Original magnification, ×4,000 (D). (E–G) Cardiac-MLCK (cMLCK) labeled with RcMK showed a diffuse cytosolic labeling pattern. (H) Cultured cardiomyocytes were stimulated with 0.2–20 μM epinephrine (Epi), which upregulated MLC2v phosphorylation in a dose-dependent manner. (I) Cultured cardiomyocytes were stimulated with 2 μM epinephrine for the indicated time periods. Epinephrine-induced phosphorylation of MLC2v in cultured cardiomyocytes was observed as early as 5 minutes after stimulation; maximal phosphorylation was obtained after approximately 30 minutes. (J–L) Cardiomyocytes cultured with serum contained organized patterns of striation and a moderate level of MLC2v phosphorylation. Middle panels show higher magnification of boxed regions in top panels. Cardiomyocytes cultured in serum-free conditions were incubated in the absence (K) or presence (L) of 2 μM epinephrine. (K) Cardiomyocytes cultured under serum-free conditions contained disorganized, punctuated actin staining with a reduced level of MLC2v phosphorylation. (L) Stimulation with epinephrine provoked rapid sarcomere reassembly and augmented MLC2v phosphorylation. Original magnification, ×1,000 (J–L, upper and lower panels); ×3,000 (J–L, middle panels).
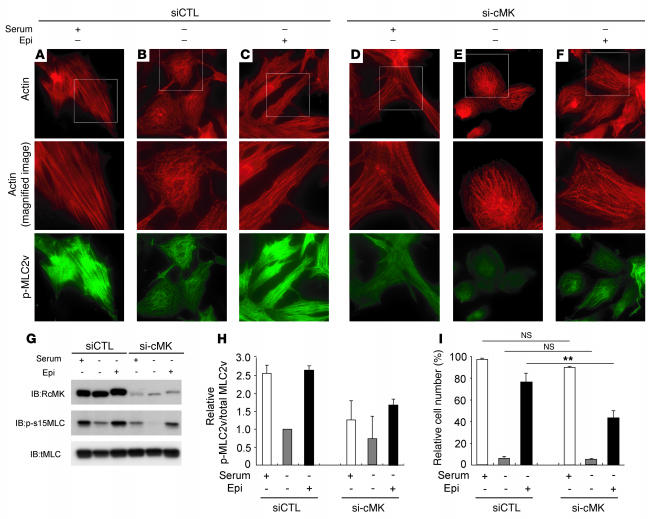
When cardiomyocytes were cultured in serum-free conditions, the organized striation pattern of actin was disrupted and the phosphorylated MLC2v-specific signal decreased (Figure 3K). To evaluate the morphologic changes observed in cardiomyocytes upon activation of endogenous cardiac-MLCK, we treated cardiomyocytes cultured under serum-free conditions with epinephrine. Stimulation of G protein–coupled receptors with epinephrine should activate cardiac-MLCK by increasing intracellular Ca2+ concentrations (13). A marked upregulation of MLC2v phosphorylation was obtained following treatment with 2 μM epinephrine (Figure 3H). Epinephrine-induced phosphorylation of MLC2v, which was observed as early as 5 minutes after stimulation, peaked within 30 minutes (Figure 3I). Treatment of the cardiomyocytes cultured in serum-free conditions with 2 μM epinephrine also induced reassembly of sarcomere structures and MLC2v phosphorylation (Figure 3, J, K, and L). To confirm the relevance of MLC2v phosphorylation by cardiac-MLCK, we introduced si-cMKs into cardiomyocytes and analyzed the sarcomere patterns in these cells. The level of phosphorylated MLC2v was reduced 72 hours after transfection with the si-cMKs; however, we did not observe any remarkable changes in the structures of the sarcomeres in cardiomyocytes cultured with serum. The sarcomeres of control siRNA– and si-cMK–treated cells contained organized filament structures (cells with organized sarcomeres, 97.0% ± 1.0% versus 90.0% ± 1.0%; NS; Figure 4, A–F and I). In contrast, the knockdown of cardiac-MLCK produced significant effects on sarcomere reassembly. si-cMK inhibited sarcomere reassembly after epinephrine treatment in cardiomyocytes cultured under serum-free conditions (cells with organized sarcomeres, 76.0% ± 8.5% versus 43.6% ± 7.0%; P < 0.005; Figure 4, A–F and I). We also confirmed the phosphorylation of MLC2v using immunoblot analysis (Figure 4G). The results of the immunoblot analysis are quantified in Figure 4H, and the relative MLC2v phosphorylation levels in this experiment exhibited a similar pattern as the percentages of cardiomyocytes with organized sarcomeres (Figure 4I), except in baseline, serum-containing conditions. These data suggest that MLC2v phosphorylation by cardiac-MLCK plays a critical role in initiating sarcomere reassembly.
Figure 4. Cardiac-MLCK regulates the initiation of sarcomere assembly in cultured cardiomyocytes through MLC2v phosphorylation.
Original magnification, ×1,000 (upper and lower panels); ×2,000 (middle panels). (A–F) Cardiomyocytes were transfected with control siRNA (A–C) or si-cMK (D–F). Middle panels show higher magnification of boxed regions in top panels. In serum-containing conditions, si-cMK–transfected cardiomyocytes showed reduced levels of MLC2v phosphorylation (D) compared with control siRNA–transfected cardiomyocytes (A), although both exhibited regularly organized sarcomere structures. Actin staining in cardiomyocytes cultured in serum-free conditions revealed a punctuated pattern in the sarcomeres (B and E); moreover, the degree of MLC2v phosphorylation was reduced in the si-cMK–transfected cardiomyocytes compared with the control siRNA–transfected cardiomyocytes. Stimulation with 2 μM epinephrine provoked upregulation of MLC2v phosphorylation and sarcomere reassembly in control siRNA–transfected cardiomyocytes (C), but not in si-cMK–transfected cardiomyocytes (F). (G) We confirmed the levels of MLC2v phosphorylation shown in A–F using immunoblot analysis. (H) Quantitation of the levels of phosphorylated MLC2v shown in G. Values are mean ± SEM. (I) Percentage of the cells with organized sarcomeres. There was no significant difference between the populations of cardiomyocytes transfected with control siRNA and si-cMK under either serum-containing or serum-free conditions. The percentage of the cells with organized sarcomeres was significantly higher for the control siRNA–transfected cardiomyocytes than for the si-cMK–transfected cardiomyocytes. Values are mean ± SEM. p-MLC2v, phosphorylated MLC2v. **P < 0.001.
Cardiac-MLCK is essential for normal cardiac development and function in zebrafish embryos.
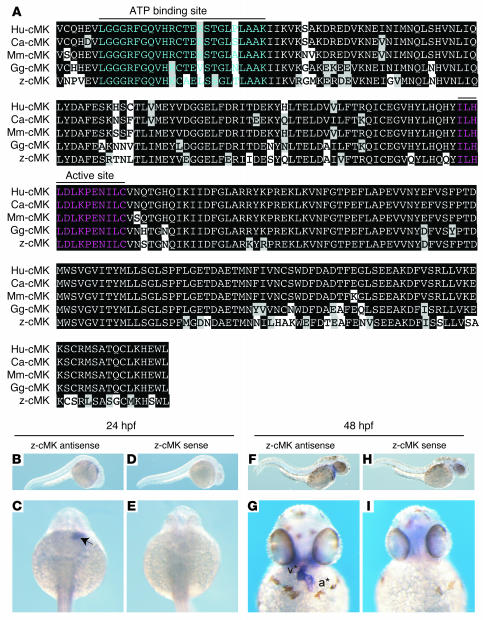
In order to further evaluate the physiologic roles of cardiac-MLCK, genetically engineered animals must be examined. In mice, however, targeted deletion of the cardiac ventricular myosin light chain, a specific substrate of cardiac-MLCK, was embryonic lethal at embryonic day 12.5 (6). Because cardiac-MLCK is an upstream modulator of MLC2v, deletion of the gene encoding cardiac-MLCK could also be embryonic lethal. Therefore, we performed in vivo knockdown experiments in Danio rerio, in which the phenotype generated by disrupting the functions of a targeted gene can be analyzed even if loss of the gene’s functions is fatal. First, we generated a zebrafish cDNA library from which we cloned the zebrafish ortholog of MYLK3 (zmylk3; encoding z–cardiac-MLCK). The amino acid sequence of cardiac-MLCK is highly similar to those of other vertebrate orthologs, especially within the C-terminal serine/threonine kinase domain (Figure 5A). Furthermore, like MYLK3, zmylk3 is located between the genes VPS35 and NP001001436.1 (Assembly Zv5sc; Wellcome Trust Sanger Institute), indicating that this was the region of synteny between human and zebrafish. We also performed whole-mount in situ hybridizations using zmylk3-specific probes; the results indicated that zmylk3 was expressed only in the heart at 24 and 48 hours postfertilization (hpf; Figure 5, B–I).
Figure 5. Cardiac-MLCK is highly conserved in several vertebrates, including zebrafish.
(A) Cardiac-MLCK is evolutionarily conserved in vertebrates, including humans (Hu), dogs (Ca), mice (Mm), chickens (Gg), and zebrafish (z), with the highest degree of homology in the C-terminal portion of the serine/threonine kinase domain. Black backgrounds indicate identical amino acids. Amino acids in the ATP-binding region are shown in blue; those in the kinase active site are shown in red. (B–I) Whole-mount in situ hybridizations depict the expression of z–cardiac-MLCK (z-cMK) in zebrafish embryos hybridized with z–cardiac-MLCK–specific antisense probe (B, C, F, and G) or z–cardiac-MLCK sense probe (D, E, H, and I). At 24 hpf, z–cardiac-MLCK was expressed in heart precursor cells (arrow). At 48 hpf, z–cardiac-MLCK was selectively expressed in the heart (asterisks denote atrium [a] and ventricle [v]).
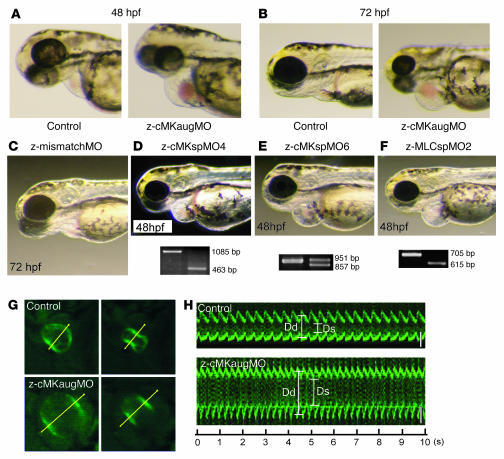
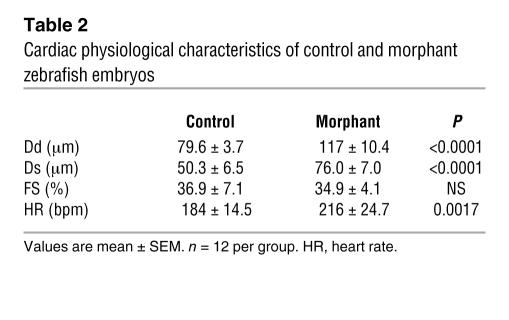
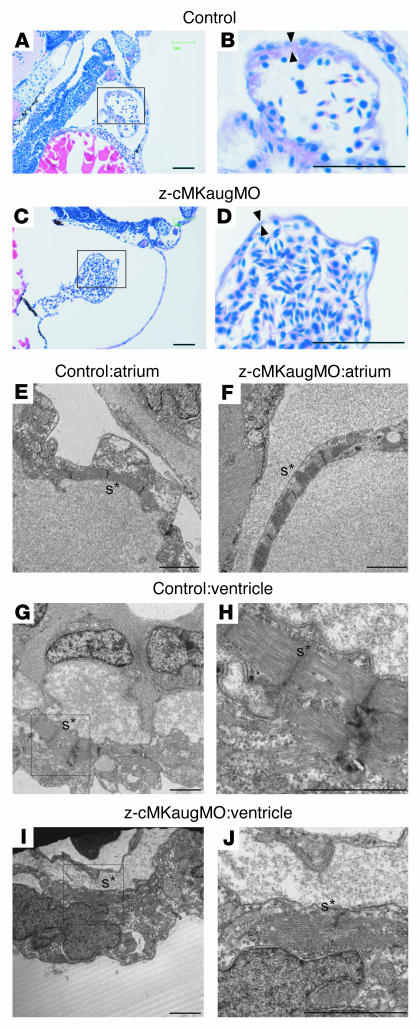
We injected zebrafish embryos with a specific MO directed against the AUG translational start site of the z–cardiac-MLCK mRNA (z-cMKaugMO). At 33 hpf, compared to control mock-injected zebrafish embryos, the heart region was slightly swollen in the z-cMKaugMO morphants. At 48 hpf, ventral swelling was observed in 45.6% ± 6.8% of the z-cMKaugMO morphants (Figure 6A). The ventral swelling became more apparent at 72 hpf (Figure 6B). In contrast, zebrafish embryos injected with an MO containing 5-base mismatches compared with z-cMKaugMO were indistinguishable from control zebrafish embryos (Figure 6C). We further examined the effects of 3 additional MOs, which were targeted to delete specific exons of z–cardiac-MLCK and z-MLC2v. Of these MOs, 2 were directed against the splice donor and acceptor sites of exons 4 and 6 of z–cardiac-MLCK, respectively. Deletion of exon 4 caused a frameshift and resulted in premature termination of the transcript. Exon 6 includes the catalytic center of z–cardiac-MLCK, and its deletion was expected to diminish the protein’s kinase activity. The third MO was designed to delete exon 2 of z-MLC2v, which includes the phosphorylatable serine. These 3 MOs effectively deleted the targeted exons, inducing comparable ventral swelling phenotypes (Figure 6, D–F). The finding that 4 different MOs produced similar results suggests that the cardiac phenotypes resulted from a loss of the kinase activity of z–cardiac-MLCK. To evaluate the cardiac phenotype of the z-cMKaugMO morphants in detail, we examined the SAG4A zebrafish strain, which specifically expresses GFP in the cardiac ventricle (14). After injecting z-cMKaugMO into SAG4A embryos, cardiac motion at 72 hpf was imaged with a high-sensitivity digital camera attached to a fluorescence stereomicroscope (Figure 6G and Supplemental Movies 1 and 2; supplemental material available online with this article; doi:10.1172/JCI30804DS1). Recordings were converted to motion mode (M-mode) images using our original software (Figure 6H). From these images, we determined the end-diastolic dimension (Dd), end-systolic dimension (Ds), and fractional shortening (FS) of the cardiac ventricle. These data are summarized in Table 2, and the results indicate that the cardiac dimensions of the z-cMKaugMO morphants were significantly larger than those of control zebrafish embryos (Dd, 79.6 ± 3.7 versus 117.0 ± 10.4 μm; Ds, 50.3 ± 6.5 versus 76.0 ± 7.0 μm; P < 0.0001 for both comparisons). We did not, however, observe a significant difference in cardiac contractility as assessed by the FS (36.9% ± 7.1% versus 34.9% ± 4.1%; NS), likely because of a compensatory upregulation of inotropy. In support of this hypothesis, we observed that the heart rate was significantly higher in the z-cMKaugMO morphants (184 ± 14.5 versus 216 ± 24.7 bpm; P = 0.0017). At 5–6 days after fertilization, the z-cMKaugMO morphants developed systemic edema and died of circulatory disturbances. Histopathologic analysis demonstrated that the ventral swelling in the z-cMKaugMO morphants reflected pericardial edema. Although the cardiac atria were almost normal, the ventricular walls of the morphants were thinner than those of control zebrafish embryos (Figure 7, A–D). Transmission electron microscopy revealed that only a few poorly differentiated sarcomere structures were present in the ventricles of the z-cMKaugMO morphants (Figure 7, G–J); no other apparent abnormalities were detected in the atrial sarcomeres (Figure 7, E and F). These data suggest that cardiac-MLCK is required for sarcomere formation in the developing heart.
Figure 6. Suppression of z–cardiac-MLCK expression induced dilatation of the cardiac ventricle in zebrafish embryos.
(A and B) Control mock-injected zebrafish embryos and zebrafish embryos injected with z-cMKaugMO produced the phenotype of ventral swelling at 48 hpf (A) and 72 hpf (B). (C) Zebrafish embryos injected with MOs with 5-base mismatch to z-cMKaugMO (z-mismatchMO) showed phenotypes comparable to those of controls. (D and E) Injection of specific MOs designed to interfere with the splicing of z–cardiac-MLCK exon 4 (z-cMKspMO4; D) or exon 6 (z-cMKspMO6; E) or with the splicing of z-MLC2v exon 2 (z-MLCspMO2; F), which coded for the phosphorylatable serine residue, also induced the phenotype of ventral swelling. RT-PCR products amplified from cDNA produced from the morphants were shorter than those obtained from control embryos due to the removal of the targeted exons. (G) Cardiac motion in the control embryos and z-cMKaugMO morphants. Shown are end-diastolic (left) and end-systolic (right) phases of the cardiac ventricular cycle in a control embryo and z-cMKaugMO morphant. (H) Representative M-mode images of both control embryo and z-cMKaugMO morphant hearts. Scale bars: 50 μm. Original magnification, ×20 (A–F); ×100 (G).
Table 2 .
Cardiac physiological characteristics of control and morphant zebrafish embryos
Figure 7. Histology of the zebrafish heart at 48 hpf.
(A–D) Longitudinal sections stained with hematoxylin and eosin. Scale bars: 50 μm. (E–J) Transmission electron micrographs. Scale bars: 2 μm. (A and B) Histology of control zebrafish hearts at 48 hpf. A relatively thick ventricular wall was apparent (B, arrowheads). (C and D) Pericardial edema and a thinner ventricular wall (D, arrowheads) were observed in z-cMKaugMO morphants. (E and F) In the atria, the sarcomere structures were well differentiated in both the control embryos and the z-cMKaugMO morphants. In the ventricles of control embryos, robust sarcomere structures were observed (G and H), whereas the ventricles of the z-cMKaugMO morphants contained sparse and immature sarcomere structures (I and J). Images in B, D, H, and J show higher magnifications of the boxed areas in A, C, G, and I, respectively. Asterisks denote sarcomere structures (s).
Cardiac-MLCK is upregulated during myofibrillogenesis and in mammalian models of heart failure.
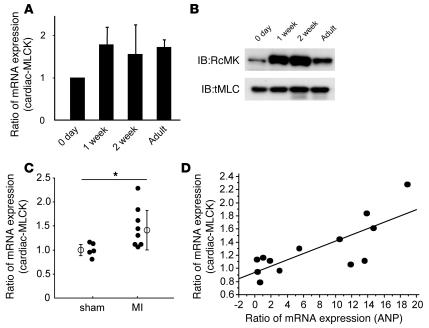
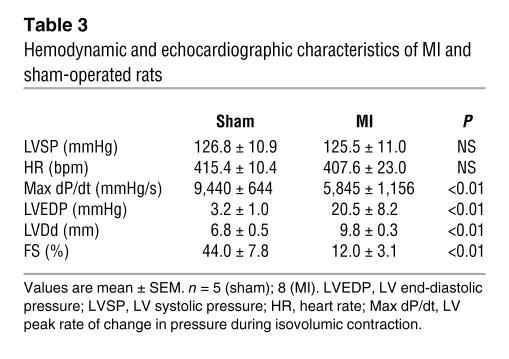
Sarcomere organization in cardiomyocytes in vivo is supposed to occur during myofibrillogenesis. In the rat heart, the mRNA and protein levels of cardiac-MLCK were upregulated from 1 week after birth through adulthood (Figure 8, A and B). The expression of cardiac-MLCK mRNA was also analyzed in mammalian models of heart failure. Myocardial infarctions (MIs) were produced in Wistar rats by permanently ligating the left anterior descending artery. At 4 weeks after the onset of MI, heart failure developed. The hemodynamic and echocardiographic parameters of the MI and sham-operated rats are summarized in Table 3. In MI rats, the LV end-diastolic pressure and LVDd were significantly higher than in sham-operated rats (LV end-diastolic pressure, 20.5 ± 8.2 versus 3.2 ± 1.0 mmHg; P < 0.01; LVDd, 9.8 ± 0.3 versus 6.8 ± 0.5 mm; P < 0.01), whereas the maximum LV peak rate of change in pressure during isovolumic contraction (Max dP/dt) and FS were significantly lower than in sham-operated rats (Max dP/dt, 5,845 ± 1,156 versus 9,440 ± 644 mmHg/s; P < 0.01; FS, 12.0 ± 3.1 versus 44.0 ± 7.8%; P < 0.01). In MI rats, MYLK3 expression was significantly upregulated compared with that in the sham-operated rats (relative cardiac-MLCK mRNA expression, 1.46 ± 0.42 versus 1.00 ± 0.15; P < 0.05; Figure 8C). Furthermore, the relative mRNA expression level of cardiac-MLCK was significantly correlated with that of ANP (r = 0.778, P < 0.005; Figure 8D). Upregulation of cardiac-MLCK expression in the infantile heart suggests cardiac-MLCK participates in myofibrillogenesis. Additionally, upregulation of cardiac-MLCK mRNA levels in mammalian models of heart failure confirmed the results obtained with the microarray analysis of human failing myocardia.
Figure 8. Expression of cardiac-MLCK is upregulated in infantile rat myocardia and failing rat myocardia.
(A) mRNA expression of cardiac-MLCK was also upregulated in rat myocardia from 1 week after birth to adulthood. The levels of cardiac-MLCK protein were upregulated in infantile myocardia 1–2 weeks after birth. (B) The levels of cardiac-MLCK protein were upregulated in infantile myocardia 1–2 weeks after birth. (C) mRNA expression of cardiac-MLCK was significantly upregulated in failing rat myocardia. n = 5 (sham-operated); 8 (MI). Filled symbols represent values from individual mice; open symbols with bars represent mean ± SEM. *P < 0.05. (D) The relative mRNA expression levels of ANP and cardiac-MLCK were significantly correlated (r = 0.778; P < 0.005).
Table 3 .
Hemodynamic and echocardiographic characteristics of MI and sham-operated rats
Discussion
In this study, we performed microarray analysis of human failing myocardia to identify new genes involved in the pathophysiology of CHF. By comparing mRNA expression analysis with the clinical parameters of the patients, we identified what we believe to be a novel candidate gene, MYLK3 (encoding cardiac-MLCK), that had not been isolated in previous microarray studies of heart failure (15). Upregulation of MYLK3 transcription in failing myocardia was confirmed in mammalian models of heart failure, such as MI rats. In this experiment, mRNA expression of cardiac-MLCK was significantly upregulated in MI rats with heart failure, and the relative expression profile was well correlated with that of ANP, a representative marker of CHF.
MLCK family members in muscle are sarcomeric protein kinases that phosphorylate a serine residue near the amino terminus of the myosin regulatory light chain. In cardiac muscle, phosphorylation of MLC2v led to sarcomere organization, an event that represents cardiac hypertrophy in cultured neonatal rat cardiomyocytes (13). skMLCK is thought to be the predominant kinase that acts on MLC2v, and a gradient of MLC2v phosphorylation in the cardiac wall from endocardium to epicardium is responsible for the generation of cardiac torsion (9). A recent study using skMLCK-deficient mice, however, revealed that removing skMLCK did not result in a cardiac phenotype (10). Furthermore, in the current study and previous studies, skMLCK expression was not detected in the heart by either Western blotting or RT-PCR (16), suggesting the existence of an as-yet unknown kinase that phosphorylates MLC2v in cardiac muscle.
We identified cardiac-MLCK, which serves as a specific kinase for MLC2v in cardiac muscle. In cultured cardiomyocytes, cardiac-MLCK regulates sarcomere assembly through the phosphorylation of MLC2v. When isolated cardiomyocytes were cultured under serum-free conditions, established sarcomere structures were disrupted. Overexpression of recombinant cardiac-MLCK and exogenous stimulation by epinephrine promoted sarcomere reassembly through MLC2v phosphorylation. Similar findings have previously been reported using recombinant constitutively active skMLCK (13). We further elucidated the physiologic roles of endogenous cardiac-MLCK using siRNAs. Decreases in MLC2v phosphorylation following the introduction of si-cMK significantly impaired epinephrine-induced sarcomere reassembly. Additionally, specific knockdown of cardiac-MLCK did not affect to the expression of other sarcomere-related proteins such as troponin T, desmin, and α-actinin. These proteins are thought to have important roles in sarcomere and myofibril formation (17–19). Thus, in cardiomyocytes, phosphorylation of MLC2v by cardiac-MLCK is an essential step for the initiation of sarcomere assembly. Upregulation of the protein levels of cardiac-MLCK in infantile rat heart supports this idea.
In this experimental model, no phenotypic alterations were observed following knockdown of cardiac-MLCK in cultured cardiomyocytes. This apparently paradoxical result occurred because phosphorylation of MLC2v is upregulated in cultured cardiomyocytes until 36 hours after plating, after which it is gradually downregulated. In the siRNA-mediated gene knockdown experiment, a reduction in the cardiac-MLCK protein level that was sufficient to decrease the phosphorylation of MLC2v was only obtained 60–72 hours after isolation. Therefore, by the time the required level of protein suppression was achieved, primary sarcomere assembly had been completed, and the subsequent decreases in MLC2v phosphorylation did not disrupt established sarcomere structures.
Reduction of cardiac-MLCK levels in zebrafish embryos through the injection of z-cMKaugMO resulted in ventral swelling, which has been previously reported to be a representative phenotype of cardiac abnormalities in zebrafish embryos (20, 21). The reliability of the results obtained with z-cMKaugMO was confirmed using multiple MOs that targeted not only cardiac-MLCK but also its substrate, MLC2v. In each experiment, reproducible results were obtained. Another MO that has 5-base mismatch to z-cMKaugMO was also examined as a negative control MO. Further analysis revealed dilatation of the ventricle with a thinned ventricular wall and immature sarcomeres in the morphants. The fragility of the ventricular wall as a result of insufficient sarcomere formation may have caused the ventricular dilatation. Although ventricular function as assessed by FS was preserved in the morphants, this might have been due to some positive inotropic effects, which were suggested by the increased heart rate observed in the z-cMKaugMO morphants. Although several reports have investigated the effects of MLC2v phosphorylation in striated muscle contractions, including in cardiac muscle, the in vivo ventricular role of MLC2v phosphorylation is still not well understood (22, 23). To explore how cardiac-MLCK contributes to ventricular function, other experiments, such as a skinned fiber study, should be performed. A similar cardiac phenotype was reported in a recent study investigating the zebrafish tel mutant, in which the gene encoding MLC2v was disrupted by an N-ethyl-N-nitrosourea-induced mutation. The authors concluded that MLC2v is essential for the assembly of myosin thick filament (24). The observation of incomplete sarcomere formation resulting in a dilated ventricle in zebrafish embryos after injection of z-cMKaugMO can be explained by an inability to initiate sarcomere assembly as a result of reduced cardiac-MLCK levels.
Our results prompt the important question of how cardiac-MLCK is involved in the pathophysiology of CHF. In failing myocardia, decreases in myofibrillar proteins such as titin, myosin, and actin, together with the sarcomere defects, have been identified (25, 26). Reduced expression of MLC2v protein as a result of protease-mediated cleavage and reduced phosphorylation of MLC2v have also been reported in the myocardia of patients with dilated cardiomyopathy. These changes produced unstable, short myofilaments following defective assembly of the myosin thick filaments (27, 28). Our preliminary data also revealed that the protein expression of cardiac-MLCK and the extent of MLC2v phosphorylation were remarkably decreased in failing myocardia of trans-aortic constriction mice compared with those of sham-operated mice. Previous reports and our present results suggest that cardiac-MLCK may be upregulated to compensate for the lower expression and reduced phosphorylation of MLC2v. As a possible therapeutic modality in patients with CHF, upregulation of cardiac-MLCK may promote sarcomere reassembly and enhanced contractility of the failing heart.
Methods
Animals.
All procedures were performed in conformity with the Guide for the care and use of laboratory animals (NIH publication no. 85-23, revised 1996) and were approved by the Osaka University Committee for Laboratory Animal Use.
Materials.
We used commercially available anti-FLAG-M2 antibody and anti-FLAG-M2 affinity gel (Sigma-Aldrich), monoclonal mouse anti-troponin T cardiac isoform antibody (NeoMarkers), monoclonal mouse anti-human desmin Antibody (Dako Corp.), and polyclonal goat anti–α-actinin (N-19) antibody (Santa Cruz Biotechnology Inc.). Epinephrine hydrochloride was purchased from Sigma-Aldrich. We also generate RcMK, anti-human smMLCK, tMLC, and p-s15MLC.
Microarray analysis.
For microarray analysis, 2 RNA samples of human normal myocardium and 12 samples of failing myocardium were used. Failing myocardium samples were obtained from severe CHF patients by Batista or Dor operation after obtaining the patients’ written informed consent. PAP was measured 2–4 weeks before the operation, and ejection fraction (EF) was measured by echocardiography the day before the operation. Normal samples were purchased from Biochain Inc. Cardiac gene expression was determined using the HG-U95 Affymetrix GeneChip. All expression data were normalized by global scaling and analyzed by GeneSpring software (Agilent Technologies). All expression data were normalized per gene and analyzed after removing noise and unreliable data. PAP, EF, and BNP values were normalized to their median values, and the correlation between gene expression and the clinical parameters was evaluated. To further select genes that are expressed almost exclusively in heart, expression values for the candidate genes were retrieved in 24 major tissues for analysis from GeneExpress database (Gene Logic Inc.) containing GeneChip expression profiles of human samples.
RNA extraction, RT-PCR, and quantification.
Rat tissues (20–50 mg) and zebrafish embryos at 72 hpf were homogenized in 1 ml RNA-Bee reagent (Tel-Test Inc.), and total RNA was isolated and converted to cDNA using Omniscript RT kit (Qiagen) according to the manufacturer’s instructions. Specific primers to amplify rat ANP, β myosin heavy chain, cardiac-MLCK, and GAPDH mRNA were purchased from Applied Biosystems. Quantitative RT-PCR reactions were run in duplicate using the ABI Prism 7700 Sequence Detector System (Applied Biosystems). The level of each transcript was quantified by the threshold cycle (Ct) method using GAPDH as an endogenous control. For RT-PCR, specific primers that cover the region of targeted exons were designed to amplify the transcripts of z–cardiac-MLCK and z-MLC2v. See Supplemental Methods for primer sequences.
Northern blot analysis.
Commercially available human multiple tissue Northern blot and polyA+ RNA of human heart and skeletal muscle were purchased from Clontech. Each polyA+ RNA was reverse transcribed and amplified using an Omniscript RT kit (Qiagen) according to the manufacturer’s protocol. Hybridization probes of human cardiac-MLCK and smMLCK were amplified by PCR from cDNA of human heart, and a hybridization probe of human skMLCK was amplified by PCR from cDNA of human skeletal muscle. Membrane was hybridized to 32P–labeled probe in Rapid–Hyb buffer (Amersham Bioscience) at 65°C for 1 hour. Final wash conditions were 0.1× SSC with 0.1% SDS at 65°C for 5 minutes. Hybridized membrane was visualized by autoradiography using the BAS system (Fuji).
Preparation and transfection of adenovirus constructs.
Adenovirus constructs were generated using ViraPower Adenoviral Expression System (Invitrogen) essentially as instructed by the manufacturer. Adenovirus vectors encoding murine cardiac-MLCK and LacZ were infected to cultured cardiomyocytes for 12 hours in various MOIs. Protein collection and immunostaining were performed 48 hours after adenovirus infection.
Identification of the substrate of cardiac-MLCK.
Recombinant cardiac-MLCK was expressed in HEK293T cells as FLAG-tagged protein. HEK293T cells expressing FLAG-tagged cardiac-MLCK were lysed with cell lysis buffer (20 mM MOPS, pH 7.0, 0.15 M NaCl, 10% glycerol, and 1% CHAPS) and recombinant cardiac-MLCK was purified by immunoprecipitation using anti-FLAG-M2 affinity gel (Sigma-Aldrich). Hearts dissected from male C57BL/6 mice (10–12 weeks of age) were mechanically homogenized using a Polytron homogenizer in 10 ml of tissue lysis buffer (30 mM MOPS, pH 6.8, 5% glycerol, 0.1% 2-mercaptoethanol, and 1 mM EGTA). Lysate was centrifuged for 40 minutes at 100,000 g, and 9 ml of supernatant was collected. Murine heart extracts were then applied to SP650 cation exchange column. The column was equilibrated with elution buffer A (30 mM MOPS, 5% glycerol, 0.1% 2-mercaptoethanol) at pH 6.8, and the extracts were eluted with a linear gradient of NaCl (0–0.5 M) at a flow rate of 1 ml/min. Each 1-ml fraction collected was incubated for 30 minutes with activated recombinant cardiac-MLCK, commercially available recombinant calmodulin (Upstate), 2 mM CaCl2, and [γ-P32]ATP and then subjected to SDS-PAGE. After drying, the gel was autoradiographed and visualized with BAS (Fuji). The fractions containing 20-kDa substrate (fractions 10 and 11) labeled with [γ-P32]ATP were pooled and applied to a phenyl-RPLC column (5Ph-AR-300; nacalai tesque) equilibrated with 0.3% trifluoroacetic acid and 5% acetoritrile. Fractions were eluted with a linear gradient of 100% acetonitrile at flow rate of 1 ml/min. After separation with SDS-PAGE, the gel was simultaneously silver stained and autoradiographed. After identifying the 20-kDa substrate with silver-stained gel, the bands were excised from the gel, and proteins were identified by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry and peptide mass fingerprinting.
Preparation of cultured neonatal rat cardiomyocytes and gene silencing via RNA interference.
Primary cultures of neonatal cardiomyocytes were prepared from Wistar rats as described previously (29). Cardiomyocytes were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Equitech-Bio). At 6 hours after isolation of cardiomyocytes, cells were transfected with siRNAs (100 nmol/l) using Optifect reagent (Invitrogen) according to the manufacturer’s instructions. Both si-cMK (see Supplemental Methods) and si-smMK (see Supplemental Methods) were purchased from B-bridge. As a negative control, cells were transfected with siControl Non-Targeting siRNA#1 (B-bridge). Isolation of mRNA was performed at 24 hours after transfection and protein experiments were performed at 72 hours after transfection. For immunostaining, the same procedures of siRNA transfection were performed in one-fifth scale on Lab-Tek Chamber Slides (nunc).
Cloning of z–cardiac-MLCK.
We generated an adult zebrafish cDNA library in Lambda Zap II (Stratagene) using polyA+ RNA from adult zebrafish. The cDNA library was screened with the probe designed to the 5′ side in the ORF of the putative zebrafish ortholog of cardiac-MLCK sequence. Positive phage clone was determined by using phage plaque screen method and single clone excision protocol according to the manufacturer’s instructions (Stratagene).
Gene accession numbers.
DDBJ accession numbers for the zebrafish MLCK family were as follows: cardiac-MLCK, AB267907; smMLCK, AB267908; skMLCK, AB267909.
Whole-mount in situ hybridization.
The digoxigenin-labeled antisense and sense RNA probes (see Supplemental Methods) were transcribed using SP6 and T7 RNA polymerase. Zebrafish embryos at 24 and 48 hpf were fixed with 4% paraformaldehyde, digested with proteinase K, and hybridized with each probe at 68°C. Alkaline-conjugated anti-digoxigenin antibody was used to detect the signals. After staining, embryos were refixed with 4% paraformaldehyde and stored in PBS.
Injection of MO.
All MOs were synthesized by Gene-Tools. At cell stages 1–4, 4–10 ng of these MOs were injected into zebrafish embryos. Several data were collected before the 96-hpf stage. Sequences of MOs are available in the Supplemental Methods.
Analysis of zebrafish cardiac histology and cardiac function.
We studied hearts of control mock-injected zebrafish embryos and z-cMKaugMO–injected zebrafish embryos at 72 hpf by routine histopathology including transmission electron micrography. To visualize the motion of zebrafish cardiac ventricle, the SAG4A strain of zebrafish, which specifically expresses GFP in its cardiac ventricular wall (14), was applied to MO-mediated gene knockdown experiments. GFP-expressed control mock-injected and z-cMKaugMO–injected zebrafish hearts at 72 hpf were imaged with Leica digital camera DFC 350 FX on a Leica MZ 16 FA fluorescence stereomicroscope. Acquired images were compiled as digital movie files using Leica FW4000 software. Each recorded movie was converted to M-mode image using our original software, and Dd, Ds, FS, and heart rate were measured from the M-mode images.
Experimental protocols of rats.
Male Wistar rats (0 days, 1 week, 2 weeks, and 10 weeks for mRNA and protein expression analysis; 8 weeks for production of MI rats; Japan Animals) were used in these experiments. MI was induced by permanent ligation of the left anterior descending coronary artery as previously described (29). The same surgical procedure was performed in a sham-operated group of rats except that the suture around the coronary artery was not tied. Isolation of total RNA was performed at 4 weeks after the onset of MI from noninfarcted myocardiums of resected LVs.
Statistics.
Statistical analysis was performed using Mann-Whitney U test and single regression analysis. Data are presented as mean ± SEM. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Ayako Hara (Core Technology Research Laboratories, Sankyo Co. Ltd.) for 5′-RACE analysis; Junichi Okutsu and Masatoshi Nishimura (Core Technology Research Laboratories, Sankyo Co. Ltd.) for microarray data analysis and critical reading of the manuscript; Tomoko Morita for technical assistance; Yulin Liao, Hidetoshi Okazaki, Hiroyuki Yamamoto, and Hisakazu Kato for thoughtful discussion; and A. Kawahara (Kyoto University) for establishing the zebrafish culture system. This study was supported by a grant from the Japan Cardiovascular Research Foundation; by Grants-in-aid for Human Genome, Tissue Engineering and Food Biotechnology (H13-Genome-011) and for Comprehensive Research on Aging and Health [H13-21 seiki (seikatsu)-23], both Health and Labour Sciences Research Grants from the Ministry of Health, Labor, and Welfare; by the Takeda Science Foundation; and by a Grant-in-aid for Scientific Research (no. 17390229) from the Ministry of Education, Science and Culture of Japan.
Footnotes
Nonstandard abbreviations used: ANP, atrial natriuretic peptide; BNP, brain natriuretic peptide; CHF, chronic heart failure; cardiac-MLCK, cardiac-specific MLCK; Dd, end-diastolic dimension; Ds, end-systolic dimension; FS, fractional shortening; hpf, hours postfertilization; MI, myocardial infarction; MLC2v, myosin regulatory light chain 2, ventricular/cardiac muscle isoform; MLCK, myosin light chain kinase; M-mode, motion mode; MO, morpholino antisense oligonucleotide; p-s15MLC, antibodies for phosphorylated MLC2v; PAP, pulmonary arterial pressure; RcMK, antibodies specific for rodent cardiac-MLCK; si-cMK, siRNA targeting cardiac-MLCK; si-smMK, siRNA targeting rat smMLCK; skMLCK, skeletal muscle MLCK; smMLCK, smooth muscle MLCK; tMLC, antibodies for total MLC2v; z-, zebrafish; z-cMKaugMO, MO targeting the AUG translational start site of z–cardiac-MLCK.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2812–2824 (2007). doi:10.1172/JCI30804
References
- 1.Jessup M., Brozena S. Heart failure. . N. Engl. J. Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.Kamisago M., et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 3.Olson T.M., Michels V.V., Thibodeau S.N., Tai Y.S., Keating M.T. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 4.Watkins H., et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 5.Collins J.H. Myoinformatics report: myosin regulatory light chain paralogs in the human genome. J. Muscle Res. Cell Motil. 2006;27:69–74. doi: 10.1007/s10974-006-9057-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., et al. Selective requirement of myosin light chain 2v in embryonic heart function. . J. Biol. Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- 7.Olsson M.C., Patel J.R., Fitzsimons D.P., Walker J.W., Moss R.L. Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am. J. Physiol. Heart Circ. Physiol. 2004;287:H2712–H2718. doi: 10.1152/ajpheart.01067.2003. [DOI] [PubMed] [Google Scholar]
- 8.Kamm K.E., Stull J.T. Dedicated myosin light chain kinases with diverse cellular functions. J. Biol. Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 9.Davis J.S., et al. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell. 2001;107:631–641. doi: 10.1016/s0092-8674(01)00586-4. [DOI] [PubMed] [Google Scholar]
- 10.Zhi G., et al. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazar V., Garcia J.G. A single human myosin light chain kinase gene (MLCK; MYLK). Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 12.Ruppel K.M., Uyeda T.Q., Spudich J.A. Role of highly conserved lysine 130 of myosin motor domain. In vivo and in vitro characterization of site specifically mutated myosin. J. Biol. Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- 13.Aoki H., Sadoshima J., Izumo S. Myosin light chain kinase mediates sarcomere organization during cardiac hypertrophy in vitro. Nat. Med. 2000;6:183–188. doi: 10.1038/72287. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami K., et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Sharma U.C., Pokharel S., Evelo C.T., Maessen J.G. A systematic review of large scale and heterogeneous gene array data in heart failure. J. Mol. Cell. Cardiol. 2005;38:425–432. doi: 10.1016/j.yjmcc.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Herring B.P., Dixon S., Gallagher P.J. Smooth muscle myosin light chain kinase expression in cardiac and skeletal muscle. Am. J. Physiol. Cell Physiol. 2000;279:C1656–C1664. doi: 10.1152/ajpcell.2000.279.5.C1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sehnert A.J., et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 18.Bar H., et al. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehler E., Rothen B.M., Hammerle S.P., Komiyama M., Perriard J.C. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J. Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- 20.Schonberger J., et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 21.Ebert A.M., et al. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17705–17710. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis J.S., Satorius C.L., Epstein N.D. Kinetic effects of myosin regulatory light chain phosphorylation on skeletal muscle contraction. Biophys. J. 2002;83:359–370. doi: 10.1016/S0006-3495(02)75175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dias F.A., et al. The effect of myosin regulatory light chain phosphorylation on the frequency-dependent regulation of cardiac function. J. Mol. Cell. Cardiol. 2006;41:330–339. doi: 10.1016/j.yjmcc.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Rottbauer W., et al. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ. Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- 25.Schaper J., et al. Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation. 1991;83:504–514. doi: 10.1161/01.cir.83.2.504. [DOI] [PubMed] [Google Scholar]
- 26.Hein S., Kostin S., Heling A., Maeno Y., Schaper J. The role of the cytoskeleton in heart failure. Cardiovasc. Res. 2000;45:273–278. doi: 10.1016/s0008-6363(99)00268-0. [DOI] [PubMed] [Google Scholar]
- 27.van der Velden J., et al. The effect of myosin light chain 2 dephosphorylation on Ca2+ -sensitivity of force is enhanced in failing human hearts. Cardiovasc. Res. 2003;57:505–514. doi: 10.1016/s0008-6363(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 28.Margossian S.S., et al. Light chain 2 profile and activity of human ventricular myosin during dilated cardiomyopathy. Identification of a causal agent for impaired myocardial function. Circulation. 1992;85:1720–1733. doi: 10.1161/01.cir.85.5.1720. [DOI] [PubMed] [Google Scholar]
- 29.Wakeno M., et al. Long-term stimulation of adenosine A2b receptors begun after myocardial infarction prevents cardiac remodeling in rats. . Circulation. 2006;114:1923–1932.. doi: 10.1161/CIRCULATIONAHA.106.630087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.