Abstract
A member of a novel family of the human major histocompatibility complex (MHC) class I genes termed MIC (MHC class I chain-related genes), MICA, has been recently identified near the HLA-B gene on the short arm of human chromosome 6. The predicted amino acid sequence of the MICA chain suggests that it folds similarly to typical class I chains and may have the capacity to bind peptides or other short ligands. Therefore, MICA is predicted to have a specialized function in antigen presentation or T cell recognition. During nucleotide sequence analyses of the MICA genomic clone, we found a triplet repeat microsatellite polymorphism of (GCT/AGC)n in the transmembrane (TM) region of the MICA gene. In 68 HLA homozygous B cell lines, 5 distinct alleles of this microsatellite sequence were detected. One of them contained an additional one base insertion that created a frameshift mutation resulting in a premature termination codon in the TM region. This particular allele may encode a soluble, secreted form of the MICA molecule. In addition, we have investigated this microsatellite polymorphism in 77 Japanese patients with Behçet disease, which is known to be associated with HLA-B51. The microsatellite allele consisting of 6 repetitions of GCT/AGC was present at significantly higher frequency in the patient group (Pc = 0.00055) than in a control population. Furthermore, the (GCT/AGC)6 allele was present in all B51 positive patients and in an additional 13 B51 negative patients. These results suggest the possibility of a primary association of Behçet disease with MICA rather than HLA-B.
HLA (human leukocyte antigen) molecules play an important role in self/nonself discrimination by presenting antigens to T cells; invading pathogenic microorganisms such as bacteria and viruses recognized as foreign antigens can be eliminated as a result of the immune response induced by HLA antigens. The HLA region, located on chromosome 6p21.3, encompasses a 4000-kb segment that has arisen through repeated gene duplication and conversion during evolution. From telomere to centromere, the HLA is divided into three subregions, class I (2 Mb), class III (1 Mb), and class II (1 Mb) (1, 2); more than 80 expressed non-HLA genes are also located in the HLA region (1, 2), although the function of most of these genes remains uncertain.
The cDNA clone of MICA, a member of MIC [major histocompatibility complex (MHC) class I chain-related genes] family was isolated from human lung fibroblasts and keratinocyte libraries (3). The MICA cDNA sequence is 1382 bp in length and has a single long ORF specifying a 383-amino acid protein with a relative molecular mass of 43 kDa. The MICA transcript was detected specifically in fibroblast and epithelial cells (3). The putative amino acid sequence of MICA shows 15–36% identity with various MHC class I chains from multiple species (3). Although this degree of similarity is substantially lower than that found among the MHC class I chains in any species, most of the matching residues are common to all of the aligned sequences among various class I chains. The predicted three-dimensional structure of MICA is similar to those of various class I antigens and some of the amino acid side chains known to interact with the termini of the short peptide bound by typical class I molecule are preserved in MICA. Thus it has been postulated that the MICA chain folds similarly to class I chains and may have the capacity to bind peptides or short ligands (3). It is peculiar that MICA has a gap in the α1 domain similar to that in the T27b chain which can be recognized by γδ T cells (4). Therefore, MICA may have adapted for some specialized function, presumably early in the evolution of MHC class I genes.
The MICA gene, spanning over 11 kb of DNA, is located about 40 kb centromeric to the HLA-B gene. Previously, we reported the complete nucleotide sequence and elucidated its exon–intron organization (5). Interestingly, a large number of diseases are known to be associated with particular alleles of the HLA-B and -C genes, such as ankylosing spondylitis (6), Reiter syndrome (7), Behçet disease (8), Kawasaki disease (9), psoriasis vulgaris (10), Salmonella arthritis (11), Yersinia arthritis (11), and many infectious diseases (12). Furthermore, the region between the HLA-B and BAT1 genes has been implicated in the development of several autoimmune diseases including Behçet disease (13), insulin-dependent diabetes mellitus (14), and myasthenia gravis (15). Because γδ T cells, which may recognize MICA binding peptides, could potentially be involved in autoimmune diseases (16, 17), it is very important to investigate a possible correlation between MICA genetic polymorphism and the development of these diseases.
Behçet disease is characterized by four major symptoms: oral aphthous ulcers, skin lesions, ocular symptoms, and genital ulcerations, and occasionally by inflammation in tissues and organs throughout the body (including the gastrointestinal tract, central nervous system, vascular system, lungs, and kidneys). Thus, Behçet disease is defined as a refractory systemic inflammatory disorder. It is well established that Behçet disease is associated with the HLA-B51 molecule that is relatively frequent, ranging from 45 to 60% in many different ethnic groups including Asian and Eurasian populations from Japan and the Middle East (18). However, it is not certain whether HLA-B51 itself or a closely linked gene is responsible for susceptibility to Behçet disease. Given its location and plausible function, the MICA gene is a possible candidate gene for the development of Behçet disease.
In this study, we describe a trinucleotide repeat (GCT/AGC) microsatellite polymorphism in the transmembrane (TM) region of MICA we first encountered during nucleotide sequence determination of the MICA gene (5). The allelic distribution of TM microsatellite repeat polymorphism in the MICA gene among 68 10th International Histocompatibility Workshop B cell lines is also presented. Finally, to investigate the possibility that MICA is responsible for determining the genetic predisposition to Behçet disease, TM microsatellite polymorphism was investigated in 77 Japanese patients with Behçet disease and compared with that of 103 unrelated healthy controls.
MATERIALS AND METHODS
Yeast Artificial Chromosome (YAC) Library Screening.
A YAC clone, Y109, was isolated from the YAC library constructed from the B cell line, CGM1 (A3, B8, Cw-, DR3, DQ2, DR52 and A29, B14, Cw-, DR7, DQ2, DR53), using HLA class I specific primers (19). Y109 spans 600 kb containing the HLA-B and -C genes. Y109 was partially digested by the restriction enzyme EcoRI and subcloned into the pWE15 cosmid vector (Stratagene). By screening this cosmid library using the human Alu-repeat sequence as a probe, 230 cosmid clones containing human genomic DNAs in their inserts were isolated and completely digested by EcoRI. Southern hybridization using these EcoRI fragments as probes allowed alignment of the cosmid clones into a contig. Finally, a cosmid clone, pM67, containing the MICA gene was identified by PCR amplification using the primers specific to the second exon (α1 domain) of MICA followed by Southern hybridization using the PCR product as a probe.
DNA Sequencing of the Cosmid Clone Containing the MICA Gene.
To determine the genomic sequence of the cosmid clone, pM67, we adopted the shotgun strategy for DNA sequencing (20, 21). Briefly, the MICA cosmid clone with a 37-kb insert was sonicated, repaired with Klenow fragment (Toyobo, Osaka), size fractionated (0.7–2.0 kb), and ligated into the blunt-end (SmaI site) dephosphorylated pUC19 plasmid vector. DNAs from randomly selected 500 recombinant pUC19 plasmid clones were prepared using the standard miniprep procedure modified to accommodate 10-ml bacterial cultures (20, 22) and sequenced using Taq DNA polymerase (Perkin–Elmer) and an automated fluorescent DNA sequencing machine (Applied Biosystems model 373A sequencer). Individual sequences were minimally edited to remove vector sequences, transmitted to a SPARC station 10 (Sun Microsystems, Mountain View, CA) on the tcp/ip protocol and assembled into contigs using the genetyx-Σ/sq software (Software Development, Tokyo).
B Cell Lines.
Sixty-eight HLA homozygous B cell lines used for the sequencing of the TM region of the MICA gene were provided by the 10th International Histocompatibility Workshop.
Subjects.
Seventy-seven Japanese patients with Behçet disease, and 103 unrelated and sex-matched Japanese healthy controls, were enrolled in this study. These patients were diagnosed according to the criteria proposed by the Japan Behçet’s Disease Research Committee at the Uveitis Clinic, Department of Ophthalmology, Yokohama City University School of Medicine or at the Department of Internal Medicine, Nanasawa Rehabilitation Hospital Comprehensive Stroke Center. Peripheral blood was collected after the details of this study were explained to every subject and consent to genetic screening was obtained.
Oligonucleotides.
For analysis of microsatellite repeat polymorphism in the TM region of the MICA gene, PCR primers flanking the TM region (MICA5F, 5′-CCTTTTTTTCAGGGAAAGTGC-3′; MICA5R, 5′-CCTTACCATCTCCAGAAACTGC-3′) were designed using the computer program for automatically selecting PCR primers, primer version 0.5 (Whitehead Institute for Biomedical Research). The MICA5R primer corresponds to the intron 4 and exon 5 boundary region and MICA5F is located in intron 5. Sequence primers are the same as those used for PCR amplification.
PCR Conditions and Product Purification.
A total of 100–200 ng of high molecular weight DNA were subjected to PCR amplification of the TM region in the MICA gene. Amplification reaction mixture contained 10 mM Tris·HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, 0.02% Nonidet P-40, 0.2 mM of each dATP, dTTP, dGTP, dCTP, and 0.1 mM of each primer. To each 100 μl PCR reaction mixture, 2 units DNA polymerase (Takara Shuzo, Shiga, Japan) was added. PCR amplification was carried out in a GeneAmp PCR system (Perkin–Elmer/Cetus). The reaction mixture was subjected to 1 cycle of denaturation at 94°C for 2 min followed immediately by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The PCR products were purified by chloroform, and the PCR primers were eliminated from the products by Micron-100 concentrator columns (Amicon). The 100- to 200-ng PCR products thus obtained were subjected to cycle sequencing.
Direct Sequencing of the TM Region of the MICA Gene in B Cell Lines.
The nucleotide sequences of the TM region of the MICA gene in the 68 B cell lines were analyzed by direct sequencing procedures after PCR amplification was carried out as described above. The 100 to 200-ng PCR products were subjected to cycle sequencing using the Applied Biosystems PRISM dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS (Perkin–Elmer/Cetus) according to the manufacture’s procedure and analyzed by an automated fluorescent DNA sequencing machine (Applied Biosystems model 377 sequencer).
Analysis of Triplet Repeat Polymorphism in the TM Region of the MICA Gene.
PCR was carried out by the same procedure as described above except for the use of the forward primer labeled at the 5′ end with 6-FAM amidite (Applied Biosystems). To determine the number of the triplet repeat in the TM region of the MICA gene, the amplified products were denatured for 5 min at 100°C, mixed with formamide containing a stop buffer, and electrophoresed on 6% polyacrylamide gels containing 8 M urea in an automated DNA sequencer (Applied Biosystems model 373A sequencer). The number of microsatellite repeat was estimated automatically using the genescan 672 software (Applied Biosystems) employing the local Southern method with a size standard marker of 350 TAMRA (Applied Biosystems) as well as the PCR products of the B cell lines that had been determined for the triplet repeat polymorphisms by nucleotide sequence determination as described above.
Statistical Analysis.
Gene frequencies and phenotype frequencies were estimated by direct counting. The significance of the distribution of alleles between the patients with Behçet disease and normal controls were tested by the chi-square (χ2) method with the continuity correction and Fisher’s exact probability test (P value test) (23). Furthermore, the P value was corrected by multiplication by the number of microsatellite alleles, 5, in the TM region of the MICA gene (Pc value). Comparison between two groups was made with 95% confidence interval to estimate statistical significance.
RESULTS
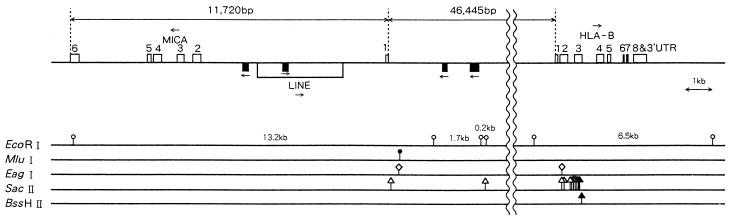
Nucleotide sequence determination of 237 kb of the chromosome 6 region including the HLA-B and -C genes in the Y109 YAC clone revealed that Y109 had the B*0801 and Cw*0701 alleles (N.M. et al., unpublished data). Therefore, the Y109 YAC clone was derived from one of the CGM1 haplotypes, A3-B8-Cw7-DR3-DQ2-DR52. The nucleotide sequence of the MICA gene associated with this haplotype was obtained by sequencing random genomic fragments (shotgun sequencing) derived from the cosmid clone, pM67, isolated by subcloning after partial EcoRI digestion of Y109. The exon–intron organization—6 exons separated by introns—was grossly similar to that of MHC class I genes (5). Our work also showed that MICA lies 46.4 kb centromeric to the HLA-B gene and that they are oriented head-to-head (Fig. 1).
Figure 1.
Genomic organization of the MICA gene and its localization within the HLA region. The MICA gene was localized 46,445 bp centromeric of the HLA-B gene and its orientation was opposite to that of the HLA-B gene, facing to each other in a head-to-head fashion. The MICA gene consisting of 6 exons separated by each intron was spanning 11,720 bp between the HLA-B and BAT1 genes. There were two Alu and one LINE (long interspersed element) sequences in intron 1. Open box on top line indicates the exon and a long open box under the top line indicates the LINE sequence. Black box corresponds to the Alu sequence. Arrow shows the orientation of genes or repetitive sequences.
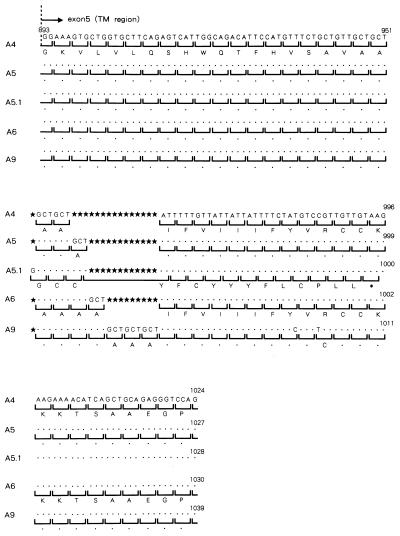
Comparing the coding sequence of the MICA gene in the pM67 cosmid clone with the MICA cDNA sequence previously reported by Bahram et al. (3), revealed a 4-nt insertion that caused a frameshift mutation resulting in a premature termination by a stop codon (TAA) in the TM region of our MICA cosmid clone (see the allele A5.1 in Fig. 2). Because this truncated MICA molecule is not rich in hydrophobic amino acid residues in the TM region, it may not reside on the cellular membrane, but rather be secreted and soluble. These facts prompted us to screen for other genetic polymorphisms in the MICA TM region using 68 HLA-homozygous 10th International Histocompatibility Workshop B cell lines. As a result, additional triplet repeat (GCT/AGC) polymorphisms were identified in the MICA TM region.
Figure 2.
Microsatellite polymorphism in the TM region (exon 5) of the MICA gene. Alphabetical character under the bracket represents amino acid abbreviation. Dot shows the same nucleotide or amino acid as upper one. Black star shows nucleotide deletion site, and ∗ indicates stop codon.
In the 68 B cell lines examined, five distinct alleles in the TM region of the MICA gene were detected. As shown in Fig. 2, the alleles consist of 4, 5, 6, and 9 repetitions of GCT/AGC (alanine) or 5 repetitions of GCT/AGC with 1 additional nucleotide insertion (G/C); they are designated A4, A5, A6, A9, and A5.1 respectively. A4 and A5.1 correspond to the alleles associated with the reported cDNA sequence (3) and the pM67 cosmid sequence from the A3-B8-Cw7 haplotype described above.
As shown in Table 1, the A5.1 allele, encoding the possibly secreted MICA molecule, was most frequently observed among the B cell lines derived from Caucasians. As expected given the location of MICA, there was a strong linkage disequilibrium between the HLA-B antigens and the MICA microsatellite alleles (Table 1). Most notable were strong associations of A4 with B18 and B27; A5 with B62; A5.1 with B7, B8, and B60; A6 with B44, B51, and B52; and A9 with B35.
Table 1.
Microsatellite polymorphism in the TM region (exon 5) of the MICA gene in B cell lines
| Cell line | B | C | A | GCT repeats | Ethnic origin | Cell line | B | C | A | GCT repeats | Ethnic origin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT8 | 7 | 7 | 3 | A5.1 | Italian | EK | 44 | 5 | 2 | A5.1 | Scandinavian |
| SCHU | 7 | 7 | 3 | A5.1 | French | SPO010 | 44 | 5 | 2 | A5.1 | Italian |
| BM14 | 7 | 7 | 3 | A5.1 | Italian | WT47 | 44 | 5 | 32 | A5.1 | Italian |
| HHKB | 7 | 7 | 3 | A5.1 | Dutch | MOU | 44 | — | 29 | A6 | Danish |
| SAVC | 7 | 7 | 3 | A5.1 | French | (MANN | 44 | — | 29 | A6 | Danish) |
| HO104 | 7 | 7 | 3 | A5.1 | French | PITOUT | 44 | — | 29 | A6 | South African Caucasoid |
| SA | 7 | 7 | 24 | A5.1 | Japanese | PF97387 | 44 | — | 29 | A6 | French |
| COX | 8 | 7 | 1 | A5.1 | South African Caucasoid | HOR | 44 | — | 33 | A6 | Japanese |
| VAVY | 8 | 7 | 1 | A5.1 | French | OMW | 45 | — | 2 | A9 | African Black |
| MGAR | 8 | 7 | 26 | A5.1 | North American Hispanic | PLH | 47 | 6 | 3 | A5.1 | Scandinavian |
| PF04015 | 8 | — | 1 | A5.1 | French | BM15 | 49 | 7 | 1 | A6 | Italian |
| CGM1 | 8, (14) | 7, (8) | 3, (29) | A5.1 | Caucasoid | MANIKA | 50 | — | 3 | A6 | South African Caucasoid |
| LBUF | 13 | 6 | 30 | A5.1 | English | BM92 | 51 | 1 | 25 | A4 | Italian |
| HO301 | 14 | 8 | 3 | A6 | French | TUBO | 51 | 7 | 2, 3 | A6 | French |
| JVM | 18 | 5 | 2 | A4 | Dutch | JHAF | 51 | 8 | 31 | A6 | English |
| L0081785 | 18 | 5 | 3 | A4 | Australian Caucasoid | LUY | 51 | — | 2 | A6 | Dutch |
| QBL | 18 | 5 | 26 | A4 | Dutch | RML | 51 | — | 2 | A6 | Warao South American Indian |
| DUCAF | 18 | 5 | 30 | A4 | French | KAS116 | 51 | — | 24 | A6 | Yugoslavian |
| 31227ABO | 18 | 7 | 2 | A4 | Italian | E4181324 | 52 | — | 1 | A6 | Australian Caucasoid |
| BTB | 27 | 1 | 2 | A4 | Scandinavian | AMAL | 53 | 4 | 28 | A9 | Indian |
| JESTHOM | 27 | 1 | 2 | A4 | Scandinavian | LKT3 | 54 | 1 | 24 | A4 | Japanese |
| HOM2 | 27 | 1 | 3 | A4 | Canadian | DBB | 57 | 6 | 2 | A9 | Amish |
| WT24 | 27 | 2 | 2 | A5.1 | Italian | MADURA | 60 | 10 | 2 | A5.1 | Scandinavian |
| FPAF | 35 | 4 | 1 | A5 | Ashkenasi Jewish | SLE005 | 60 | 10 | 2 | A5.1 | American Caucasoid |
| BM9 | 35 | 4 | 2 | A9 | Italian | DKB | 60 | 10 | 24 | A5.1 | Dutch |
| WT100BIS | 35 | 4 | 11 | A9 | Italian | PE117 | 60 | 10 | 24 | A5.1 | North American Indian |
| DEU | 35 | 4 | 31 | A9 | Dutch | SWEIG007 | 61 | 2 | 29 | A5 | American Caucasoid |
| KOSE | 35 | — | 2 | A9 | German | S. PACHEO | 62 | 1 | 31 | A5 | Unknown |
| DHIF | 38 | 5 | 31 | A5.1 | English | CB6B | 62 | 9 | 1 | A5 | Australian Caucasoid |
| WDV | 38 | — | 2 | A9 | Dutch | MLF | 62 | 9 | 2 | A5 | English |
| YAR | 38 | — | 26 | A9 | Ashkenasi Jewish | BSM | 62 | 9 | 2 | A5 | Dutch |
| TEM | 38 | — | 26 | A9 | Jewish | BOLETH | 62 | 10 | 2 | A5 | Swedish |
| BM21 | 41 | — | 1 | A6 | German | OLGA | 62 | 1, 10 | 31 | A5 | American Indian |
| RSH | 42 | 2 | 68, 30 | A6 | African Black | WT51 | 65 | 8 | 23 | A5 | Italian |
In the Japanese population, A5 was most common, with a frequency of 31.6% (control in Table 2). In contrast, the A5.1 allele harboring the premature termination codon was less common and observed in only 9.2% of the Japanese population (Table 2), noticeably lower than its high frequency in Caucasian B cell lines (Table 1).
Table 2.
Gene frequencies of the microsatellite polymorphism in the TM region (exon 5) of the MICA gene in Behçet disease
| Microsatellite allele | Amplified product, bp | Control (n = 206), (%) | Patient (n = 154), (%) | χ2 | P value | R.R. |
|---|---|---|---|---|---|---|
| A4 | 179 | 35 (17.0) | 17 (11.0) | |||
| A5 | 182 | 65 (31.6) | 39 (25.3) | |||
| A5.1 | 183 | 19 (9.2) | 6 (3.9) | |||
| A6 | 185 | 53 (25.7) | 71 (46.1) | 16.203 | 0.00006 | 2.47 |
| A9 | 194 | 34 (16.5) | 21 (13.6) |
To address the possibility that the MICA gene, rather than HLA-B51, is responsible for determining the genetic predisposition to Behçet disease, triplet repeat polymorphism in the TM region of the MICA gene was investigated in 77 Japanese patients with Behçet disease and 103 controls. As listed in Table 2, the gene frequency of the A6 allele was significantly increased in the patient group, as compared with the control group (P = 0.00006, Pc = 0.00030). To further evaluate the association of the A6 allele with Behçet disease, the phenotype frequencies were investigated in the patient and control groups. As seen in Table 3, the A6 allele was observed at a phenotype frequency of as high as 74.0% in the patient group. Therefore, the phenotype frequency of the A6 allele was significantly increased in the patients with Behçet disease as compared with the healthy controls (P = 0.00011, Pc = 0.00055).
Table 3.
Phenotype frequencies of the microsatellite polymorphism in the TM region (exon 5) of the MICA gene in Behçet disease
| Microsatellite allele | Amplified product, bp | Control (n = 103), (%) | Patient (n = 77), (%) | χ2 | P value | R.R. |
|---|---|---|---|---|---|---|
| A4 | 179 | 31 (30.0) | 15 (19.5) | |||
| A5 | 182 | 54 (52.4) | 30 (39.0) | |||
| A5.1 | 183 | 17 (16.5) | 6 (7.8) | |||
| A6 | 185 | 47 (45.6) | 57 (74.0) | 14.562 | 0.00011 | 3.40 |
| A9 | 194 | 32 (31.1) | 20 (26.0) |
DISCUSSION
Although the MICA gene product has not yet been reported at the protein level and its function remains unknown, MICA encodes a molecule similar to MHC class I antigens and may share the capacity to bind peptides or other short ligands. Therefore, MICA along with MICB (24) are thought to represent a second lineage of MHC antigens and to possibly play a specialized or modified role in the immune response (3).
In this study we have found trinucleotide repeat (GCT/AGC) microsatellite polymorphism in the MICA TM region. A microsatellite consists of repetitive sequences of 2 or 3 bases; all eukaryotic DNA contains families of such repetitive sequences. The number of repetitive sequences varies among individuals, and hence microsatellites can be used as informative polymorphic markers for genetic mapping and for disease susceptibility analysis. Although microsatellites are not normally identified inside genes, it is well known that several human hereditary disease associated genes, including myotonic dystrophy (25, 26), Kennedy disease (27), spinocerebellar ataxia type I (28), Huntington disease (29), and dentatorubral-pallidoluysian atrophy (30, 31) contain CAG/CTG triplet repeats. Four of these genes contain CAG/CTG which encodes polyglutamine repeats in their respective ORFs. The development of these diseases is associated with the massive expansion of CAG/CTG repeats, whereby the severity of the disease is increased and the age of onset is lowered in successive generations. In this context, it is of great interest to explore the possibility that any microsatellite polymorphism existing in the TM region of the MICA leads to the development of hereditary diseases.
The TM region contains a majority of hydrophobic amino acids and it plays a role in anchoring the protein into the cell membrane. In this study, we have identified five microsatellite alleles in the MICA TM region. Four of them consist of 4, 5, 6, and 9 repetitions of GCT/AGC. According to the putative ORF of the MICA cDNA, the microsatellite encodes polyalanine and, therefore, the number of alanine residues differs by the number of triplet repeats. However, one allele (the A5.1 allele) contains five triplet repeats plus one additional nucleotide insertion (GGCT/AGCC), causing a frameshift mutation resulting in premature termination by the stop codon (TAA) in the TM region. As noted above, this allele may encode a secreted, soluble MICA molecule. Because MICA is specifically expressed in fibroblasts and epithelial cells (3), it might play a specialized role in immune response. Given these observations, it is very important to investigate any possible correlation between the microsatellite polymorphism in the TM region of the MICA gene and the development of several HLA-B or -C associated diseases, including Behçet disease, ankylosing spondylitis, Reiter syndrome, Kawasaki disease, psoriasis vulgaris, Salmonella arthritis, Yersinia arthritis, as well as many infectious diseases (6–13).
To initiate such analyses, we have investigated the allelic distribution of microsatellite polymorphism in the TM region of the MICA gene among Japanese patients with Behçet disease. The frequency of the A6 allele was remarkably higher in the patient group than in the control group. Forty-four out of 77 patients with Behçet disease were B51 positive (57.1%), and all of them possessed the A6 allele. Furthermore, 13 B51 negative patients possessed the A6 allele and, in total, 57 of 77 patients carried A6 (74.0%) (Pc = 0.00055), indicating that the association of the A6 allele in the MICA gene with Behçet disease is stronger than that of HLA-B51 (Pc < 0.001) (8). On the other hand, we noted that the A5.1 allele is less common in the Japanese population and that none of the patients with Behçet disease is homozygous for the A5.1 allele, whereas the A5.1 allele is frequently observed among the B cell lines originated from Caucasians. A strong association of Behçet disease with HLA-B51 has been reported in some Caucasoid populations such as Italian, Greek, French, and English (18). However, the number of patients in Caucasoid populations is remarkably lower than those in Japanese and other Asian or Middle East ethnics. This might be explained by the apparently high frequency of the A5.1 allele in Caucasoids, although the frequencies of these microsatellite alleles have not been investigated in detail in any Caucasian populations. As many as 16 alleles have been recognized in the peptide binding regions (exons 2 and 3) of the MICA gene (32). Therefore, the conspicuously high frequency of the A6 microsatellite allele in Behçet disease patients may reflect linkage disequilibrium with a certain allelic form of the MICA in the peptide binding region.
In a previous paper, we investigated the Tau-a microsatellite polymorphism located between the HLA-B and tumor necrosis factor (TNF) genes and suggested that the pathogenic gene responsible for the development of Behçet disease is not the HLA-B (HLA-B51) gene itself but rather other gene(s) between the HLA-B and TNF genes (13), precisely where MICA gene is localized. Although the immunopathogenic mechanism underlying the development of Behçet disease remains uncertain, its various characteristic symptoms have been attributed to neutrophil hyperactivity and abnormality or hyperactivity of γδ T cells (33, 34), which possibly recognize MICA-bound ligands. Obstructive angiitis in organs such as eyes (choroid and retina), oral mucosa, genitals, and the skin, all of which are likely to be exposed to infectious microorganisms, is the first step in the inflammation characteristic of Behçet disease. The MICA gene is expressed specifically in fibroblast and epithelial cells (3), again precisely where the inflammation of Behçet disease exclusively takes place. Based on the chromosomal localization of the MICA gene, its predicted function, and the strong association of the microsatellite allele, A6, in the TM region, the MICA gene is one of strong candidates for susceptibility to Behçet disease. In this respect, investigation of the expression of the protein in patients using MICA antibodies is of great importance and is now underway in our laboratory.
In conclusion, we have found trinucleotide repeat microsatellite polymorphism in the TM region of the MICA gene and document the possibility that a secreted and soluble MICA molecule exists as one of the resulting allelic forms. Importantly, one particular allele (A6) of this microsatellite was present at significantly higher frequency in patients with Behçet disease than in controls. In addition, a significant fraction of B51 negative patients were positive for the MICA A6 allele. This defines MICA as a possible candidate gene for the Behçet disease and, at the very least, questions the designation of HLA-B as the sole MHC linked pathogenic element involved in the development of the Behçet disease.
Acknowledgments
We wish to express our appreciation to Drs. Goro Inaba and Mami Ishihara who have kindly provided some peripheral blood cell samples from patients with Behçet disease and to Mr. Shoji Miyata and Miss Megumi Sato for their technical support. This work was supported by the Japan Information Center of Science and Technology, an arm of the Science and Technology Agency, Grants-in-Aid from the Ministry of Education, Science, Sports and Culture, Japan (07041166 and 08457466), a grant from the Ministry of Health and Welfare, Japan, and a research grant from the Kanagawa Academy of Science and Technology.
Footnotes
References
- 1.Campbell R D, Trowsdale J. Immunol Today. 1993;14:349–352. doi: 10.1016/0167-5699(93)90234-C. [DOI] [PubMed] [Google Scholar]
- 2.Campbell R D. EFI Newsl. 1995;13:4–9. [Google Scholar]
- 3.Bahram S, Bresnahan M, Geraghty D E, Spies T. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito K, Van Kaer L, Bonneville M, Hsu S, Murphy D B, Tonegawa S. Cell. 1990;62:549–561. doi: 10.1016/0092-8674(90)90019-b. [DOI] [PubMed] [Google Scholar]
- 5.Bahram S, Mizuki N, Inoko H, Spies T. Immunogenetics. 1996;44:80–81. doi: 10.1007/BF02602661. [DOI] [PubMed] [Google Scholar]
- 6.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 85–100. [Google Scholar]
- 7.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 107–111. [Google Scholar]
- 8.Mizuki N, Inoko H, Mizuki N, Tanaka H, Kera J, Tsuji K, Ohno S. Invest Ophthalmol Visual Sci. 1992;33:3332–3340. [PubMed] [Google Scholar]
- 9.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 145–146. [Google Scholar]
- 10.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 112–127. [Google Scholar]
- 11.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 80–83. [Google Scholar]
- 12.Tiwari J L, Terasaki P I. In: HLA and Disease Associations. Tiwari J L, Terasaki P I, editors. New York: Springer; 1985. pp. 382–401. [Google Scholar]
- 13.Mizuki N, Ohno S, Sato T, Ishihara M, Miyata S, Nakamura S, Naruse T, Mizuki H, Tsuji K, Inoko H. Hum Immunol. 1995;43:129–135. doi: 10.1016/0198-8859(94)00159-n. [DOI] [PubMed] [Google Scholar]
- 14.Delgi-Esposti M A, Abraham L J, McCann V, Spies T, Christiansen F T, Dawkins R L. Immunogenetics. 1992;36:345–356. doi: 10.1007/BF00218041. [DOI] [PubMed] [Google Scholar]
- 15.Delgi-Esposti M A, Andreas A, Christiansen F T, Schalke B, Albert E, Dawkins R L. Immunogenetics. 1992;35:355–364. doi: 10.1007/BF00179791. [DOI] [PubMed] [Google Scholar]
- 16.Holoshitz J. Res Immunol. 1990;141:651–657. doi: 10.1016/0923-2494(90)90076-b. [DOI] [PubMed] [Google Scholar]
- 17.Hounsell, E. F. & Davies, M. J. (1993) Ann. Rheum. Dis. 52, Suppl., S22–S29. [DOI] [PMC free article] [PubMed]
- 18.Ohno S, Ohguchi M, Hirose S, Matsuda H, Wakisaka A, Aizawa M. Arch Ophthalmol. 1982;100:1455–1458. doi: 10.1001/archopht.1982.01030040433013. [DOI] [PubMed] [Google Scholar]
- 19.Imai O, Olson M V. Genomics. 1990;8:297–303. doi: 10.1016/0888-7543(90)90285-3. [DOI] [PubMed] [Google Scholar]
- 20.Wilson R K, Koop B F, Chen C, Halloran N, Sciammis R, Hood L. Genomics. 1992;13:1198–1208. doi: 10.1016/0888-7543(92)90038-t. [DOI] [PubMed] [Google Scholar]
- 21.Koop B F, Rowen L, Wang K, Kuo C L, Seto D, Lenstra J A, Howard S, Shan W, Deshpande P, Hood L. Genomics. 1994;19:478–493. doi: 10.1006/geno.1994.1097. [DOI] [PubMed] [Google Scholar]
- 22.Koop B F, Rowen L, Chen W-Q, Deshpande P, Lee H, Hood L. BioTechniques. 1993;14:442–447. [PubMed] [Google Scholar]
- 23.Thompson G A. Theor Popul Biol. 1981;20:160–206. doi: 10.1016/0040-5809(81)90009-5. [DOI] [PubMed] [Google Scholar]
- 24.Bahram S, Spies T. Immunogenetics. 1996;43:230–233. doi: 10.1007/BF00587305. [DOI] [PubMed] [Google Scholar]
- 25.Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, et al. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 26.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O’Hoy K, Leblond S, Earle-Macdonald J, de Jong P J, Wieringa B, Korneluk R G. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 27.La Spada A R, Wilson E M, Lubahn D B, Harding A E, Fischbeck K H. Nature (London) 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 28.Orr H T, Chung M, Banfi S, Kwiatkowski Jr T J, Servadio A, Beaudet A L, McCall A E, Duvick L A, Ranum L P W, Zoghbi H Y. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 29.The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 30.Koide R, Ikeuchi T, Onodera O, Tanaka H, Igarashi S, Endo K, Takahashi H, Kondo R, Ishikawa A, Hayashi T, Saito M, Tomoda A, Miike T, Naito H, Ikuta F, Tsuji S. Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- 31.Nagafuchi S, Yanagisawa H, Sato K, Shirayama T, Ohsaki E, et al. Nat Genet. 1994;6:14–18. doi: 10.1038/ng0194-14. [DOI] [PubMed] [Google Scholar]
- 32.Fodil N, Laloux L, Wanner V, Pellet P, Hauptmann G, Mizuki N, Inoko H, Spies T, Theodorou I, Bahram S. Immunogenetics. 1996;44:351–357. doi: 10.1007/BF02602779. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita N, Mochizuki M, Mizushima Y, Sakane T. In: Behçet’s Disease. Wechsler B, Godeau P, editors. Amsterdam: Excerpta Medica; 1993. pp. 7–12. [Google Scholar]
- 34.Koide, T. & Abe, T. (1993) Arthritis Rheum. 36, Suppl, s206 (abstr.).