Abstract
The haploid Tetrahymena thermophila genome contains a single α-tubulin (ATU) gene. Using biolistic transformation, we disrupted one of the two copies of the ATU gene in the diploid germ-line micronucleus. The heterozygous germ-line transformants were made homozygous in the micronucleus by mating to a star strain containing a defective micronucleus. This mating, known as round 1 genomic exclusion, resulted in two heterokaryon clones of different mating types which have both copies of the ATU gene knocked out in the micronucleus but only wild-type genes in the polycopy somatic macronucleus. When these heterokaryons were mated, the exconjugant progeny cells did not grow because the new somatic macronuclei do not have any α-tubulin genes. However, when these conjugants were transformed with a functional marked ATU gene, viable transformants were obtained that contained the transforming ATU gene at the homologous locus in the new macronucleus. The exconjugant progeny could be rescued at a high efficiency (900 transformants per μg of DNA) with a wild-type ATU gene. Unlike previous macronuclear transformation protocols, this strategy should allow introduction of highly disadvantageous (but viable) mutations into Tetrahymena, providing a powerful tool for molecular and functional studies of essential genes. These knockout heterokaryons were used to demonstrate that gene transfer from somatic macronuclei to germ-line micronuclei occurs rarely if at all.
Keywords: biolistic transformation, conjugation, paromomycin
We are developing the ciliate Tetrahymena thermophila as a model for the study of microtubule functions in vivo. Tetrahymena cells maintain extremely diverse microtubule systems, but express only one type each of α- and β-tubulin proteins encoded by a single α and two β genes (1). Tetrahymena tubulins possess most of the evolutionarily conserved post-translational tubulin modifications which have been suggested to play a role in the regulation of microtubule assembly and function (refs. 2–7 and J. Gaertig and M.A.G., unpublished results). Furthermore, we have developed methods for mass transformation which, coupled with a high frequency of homologous recombination, allow replacement of all the tubulin genes in the somatic macronucleus (mac) (8–11).
In our initial studies we introduced a drug-resistant α-tubulin gene containing mutated sites of conserved secondary modifications into Tetrahymena macs containing a mutant, drug-sensitive, α-tubulin gene. In some cases, we obtained transformants with the desired mutations and were able to examine their phenotypes (ref. 11; unpublished results). However, in several cases, we could not detect the mutations of interest, although the drug-resistance marker was detected in the genome of the transformants (B.H., J. Gaertig, and M.A.G., unpublished results). The simplest explanations for these results are that the mutation either is a dominant lethal or results in disadvantageous growth in Tetrahymena. During DNA-mediated mass transformation of the polyploid mac, transforming DNA is integrated into transformants through homologous recombination (10). Thus it is possible that the initial recombination event separates the mutation under study from the selectable marker. Alternatively, since the mac contains about 45 copies of each gene, a subsequent interchromosomal recombination event between the transformed locus and a wild-type gene could separate the mutation from the selectable marker. If the mutation is deleterious for growth, continued drug selection coupled with the random segregation of mac genes (phenotypic assortment) would result in cells containing only the drug-resistance marker. This type of somatic recombination occurs frequently in Tetrahymena (refs. 10–12; B.H., L. Yu, J. Gaertig, L. Gu, and M.A.G., unpublished results). Consequently, existing techniques for transforming the somatic mac cannot distinguish dominant lethals from mutations that reduce growth and may fail to identify such mutations.
Using a recently developed method for germ-line transformation coupled with phenotypic assortment and * (star) strains having defective micronuclei (mics) (14–16) we have developed a strategy to solve this problem. To prevent interchromosomal recombination, we eliminated the wild-type α-tubulin genes by constructing heterokaryons that contain germ-line α-tubulin knockouts whose transcriptionally inert mics are homozygous for an α-tubulin gene disrupted by a neo gene (conferring paromomycin resistance) but have only drug-sensitive wild-type alleles in the transcriptionally active macs. When these paromomycin-sensitive heterokaryons conjugate, the old drug-sensitive mac is replaced by products produced by meiosis, fertilization, and mitotic divisions of the mic. As a result, the disrupted drug-resistance allele should be expressed in the new mac, which is derived from mics containing only disrupted α-tubulin loci expressing the neo genes, allowing simple drug selection for successful mating. Cells that complete this mating should be unable to grow because the new mac does not contain any functional α-tubulin genes. If the conjugating heterokaryons are transformed with an α-tubulin gene, any progeny that survive in the drug should be transformed by the exogenous gene, expressing it as the only α-tubulin gene in their mac.
Here, we describe creation of heterokaryons homozygous for germ-line knockouts of the essential α-tubulin gene and demonstrate that the strategy works as expected. This approach should be applicable to any essential gene and should greatly facilitate further mutagenic studies of the gene. Furthermore, the fact that progeny of these heterokaryons are not viable but can be rescued by exogenous α-tubulin genes argues that gene transfer from somatic macs to germ-line mics of vegetative cells or early conjugants or from old to new macs during conjugation occurs rarely, if at all.
MATERIALS AND METHODS
Strains, Culture, and Conjugation.
T. thermophila strains CU428 {Mpr/Mpr, Chx+/Chx+ [6-methylpurine-sensitive (mp-s), paromomycin-sensitive (pm-s), cycloheximide-sensitive (cy-s), VII]}, B2086 {Mpr+/Mpr+, Chx+/Chx+ (mp-s, pm-s, cy-s, II)}, and CU427 {Mpr+/Mpr+, Chx/Chx (mp-s, pm-s, cy-s, VI)} were obtained from P. J. Bruns (Cornell University, Ithaca, NY). Strains AKO2 {Mpr+/Mpr, ATU/ΔATU, Chx+/Chx+ [mp-resistant (mp-r), pm-s, cy-s, mating type unknown]} and AKO5 {Mpr+/Mpr, ATU/ΔATU, Chx+/Chx+ (mp-r, pm-s, cy-s, VII)} are G1 progeny of a cross between CU428 and B2086 that had been biolistically transformed to pm resistance in the germ-line mic with the pTUB-KO knockout construct and then assorted to pm sensitivity in the mac. Strains AAKO2 {Mpr+/Mpr+ or Mpr/Mpr, ΔATU/ΔATU, Chx+/Chx+ (mp-r, pm-s, cy-s, mating type unknown)} and AAKO5 {Mpr+/Mpr+ or Mpr/Mpr, ΔATU/ΔATU, Chx+/Chx+ (mp-r, pm-s, cy-s, VII)} are nonstar-side exconjugants from round I genomic exclusion crosses of AKO2 and AKO5 respectively mated to the star strain B*(VI), also kindly provided by P. J. Bruns. Cells were grown in SPP (17) at 30°C with shaking.
For conjugation, two strains of different mating types were washed, starved (16–24 hr, 30°C) and mated in 10 mM Tris·HCl (pH 7.5). Cells (2 × 105 per ml) were maintained in flasks at a fluid depth of about 1 cm at 30°C without agitation. Conjugation efficiency was measured as the percentage of cells in pairs 4 hr after mixing.
Transformation Vectors.
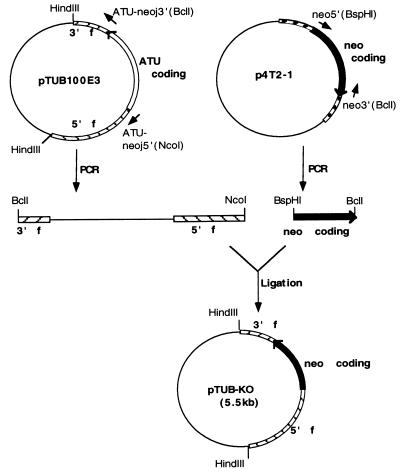
pTUB100E3-PvuII is a pBluescript KS(+) plasmid (Stratagene) containing the 3.2-kb HindIII fragment of the α-tubulin (ATU) gene of T. thermophila (18) with silent marker HaeIII and PvuII sites (see Fig. 2). To construct pTUB-KO (Fig. 1), the pBluescript vector sequence and 5′ and 3′ flanking sequences of the ATU gene were PCR amplified from pTUB100E3, a plasmid like pTUB100E3-PvuII but lacking the PvuII site. Primers were ATU-neoj5′ (NcoI) (5′-GCTTGCCATGGCTAACTTTTGATTTGGTT-3′) and ATU-neoj3′ (BclI) (5′-GTTCTTCTGATCAGTTTGATTCCTTACCTA-3′). The neomycin gene (neo) coding sequence was PCR-amplified from p4T2–1 (10) with primers neo5′ (BspHI) (5′-GTTAGTCATGACAAGCTTGGATGG-3′) and neo3′ (BclI) (5′-ATCAAACTGATCAGAAGAACTCGTCAAG-3′). The PCR products were digested respectively with the restriction enzymes indicated in the primer names and ligated to create pTUB-KO with the ATU coding sequences replaced by neo coding sequences.
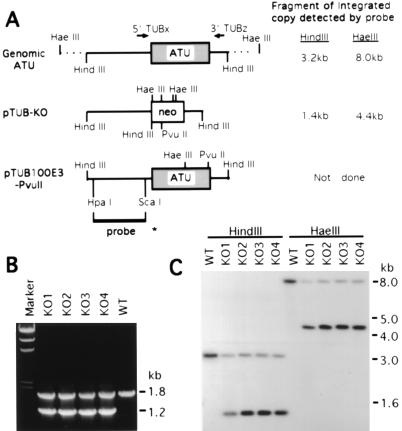
Figure 2.
(A) Maps of the macronuclear ATU gene and the transforming constructs, pTUB-KO and pTUB100E3-PvuII. The 1.4-kb ATU coding region is shown as a shaded box in an 8.0-kb HaeIII genomic fragment. In the knockout construct pTUB-KO, the 0.8-kb neo coding region (shown as an open box) contains four HaeIII sites, a HindIII site, and a PvuII site. pTUB100E3-PvuII contains a 3.2-kb cloned HindIII fragment of the ATU gene which is silently marked by two restriction sites, HaeIII and PvuII. The HpaI/ScaI fragment used as an ATU probe in the Southern blot analysis is shown as a bold line with an asterisk at its end. Fragment sizes of integrated genomic copies produced by digestion with HindIII or HaeIII which hybridize to the 5′ ATU probe are indicated, as are the sites of the PCR primers used in B. (B) PCR analysis of the ATU gene in four pTUB-KO somatic transformants (KO1–4). 5′TUBx and 3′TUBz (locations shown in A) were used as primers in PCR. The 1.8-kb band detected in wild type (WT) and in KO1–4 transformants is derived from the endogenous wild-type ATU gene. The 1.2-kb band is the size expected for the replacement of the wild-type gene by the knockout construct containing the neo gene. Marker, HindIII-cut λ DNA. (C) Southern blot analysis of the pTUB-KO transformants. Digestion with HaeIII is used to assay for homologous recombination, while HindIII digestions served as controls. When genomic DNA is digested with HaeIII, the 8.0-kb fragment derived from the wild-type ATU locus is detected in wild-type control (WT) as well as in KO1–4; the 4.4-kb band corresponds to the same locus when the neo gene replaces the ATU coding region. When digested with HindIII, the 3.2-kb band derived from the wild-type ATU gene is detected in all clones; the 1.4-kb band corresponds to the replacement fragment containing the neo gene.
Figure 1.
Construction of the α-tubulin gene (ATU) knockout plasmid. All plasmids are pBluescript-based constructs. pTUB100E3 contains a cloned HindIII fragment of the ATU gene that is marked by a silent HaeIII site; p4T2–1 contains the neo gene coding sequence; in pTUB-KO (the ATU gene knockout construct), the coding region of ATU is replaced by the coding region of neo. See text for description.
For Tetrahymena transformation, plasmids were linearized (pTUB100E3-PvuII with HindIII and pTUB-KO with EcoRI and XhoI), extracted once each with phenol/chloroform and then chloroform, precipitated with NaCl (0.2 M) and 2-propanol, and resuspended in TE (10 mM Tris·HCl, pH 8.0/1 mM EDTA) at 2 μg/μl. About 4 μg of DNA was used for each transformation.
Tetrahymena Transformation.
For biolistic transformation of the somatic mac (14), CU428 cells starved overnight in 10 mM Tris·HCl (pH 7.5), or unfed exconjugants from the cross between AAKO2 and AAKO5, were centrifuged and resuspended in the same Tris buffer at 1 × 107 cells per ml. Cells (1 × 107) were spread on a moist filter paper and bombarded with DNA-coated gold particles (1.0 μm) at 900 psi, using the DuPont Biolistic PDS-1000/He particle delivery system (Bio-Rad). Following bombardment, cells were immediately resuspended in 50 ml of SPP, incubated at 30°C for 0.5–2 hr, and then plated in 96-well microtiter plates. Two to 6 hr after bombardment, pm was added to the cells at a final concentration of 120 μg/ml.
For biolistic germ-line transformation (14), CU428 and B2086 cells were starved and mixed to initiate conjugation. At 3, 3.5, 4, and 4.5 hr after mixing, an aliquot of cells was washed with 10 mM Hepes (pH 7.5), resuspended in Hepes at 1 × 107 cells per ml, bombarded, and then resuspended in SPP and plated as described above for somatic transformation. pm (120 μg/ml) was added to the cells at 20–22 hr after bombardment. Four days later, cells were replica plated into 15 μg/ml mp to test for effective mating.
Extraction and Analysis of Genomic DNA.
For Southern blots, pTUB-KO somatic transformants (KO1–4) were grown to stationary phase in 25 ml of SPP containing 1 mg/ml pm. Genomic DNA was extracted (9), digested with HindIII or HaeIII, and blotted as described (19). Hybridization was carried out at 65°C with a 32P-labeled HpaI/ScaI fragment of the 5′ ATU flanking sequence. For analysis of the ATU gene by genomic PCR, somatic pTUB-KO transformants were grown in 1.5 ml of SPP containing 1 mg/ml pm to stationary phase, while transformed progeny of knockout heterokaryons were grown in 1.5 ml of SPP containing 100 μg/ml pm. The ATU gene in genomic DNA was amplified with the following primers (Fig. 2A): 5′TUBx (5′-CTCTTAAGCAGTCCCTCAAGT-3′) and 3′TUBz (5′-CCTTTGTATTTCTTAGTCAAGAAAGC-3′). PCR products digested with HaeIII or PvuII were analyzed by electrophoresis in 0.8% agarose gels.
Test of Gene Transfer from Soma to Germ Line and Among macs.
To cross AAKO2 × AAKO5, 100 ml each of the two strains were starved overnight and mixed to initiate conjugation. For crosses B2086 × CU428, B2086 × AAKO2, and B2086 × AAKO5, 50-ml portions of each of the two strains in each cross were starved and mixed. Twenty-one hours after mixing, cells were refed with an equal volume of 2× SPP and three 100-μl cell samples were taken and stained with 4′,6-diamidino-2-phenylindole (DAPI) (11). The numbers of unmated cells (one mac) and of successful conjugants (two or three macs; ref. 20) were counted to determine the conjugation efficiency. Three hours later, pm (120 μg/ml) was added and cells were plated with or without dilution with SPP containing 120 μg/ml pm. Wells containing growing cells were scored 4 days later. The number of pm-resistant (pm-r) cells was calculated by using the Poisson distribution (21).
RESULTS
ATU Is an Essential Gene That Can Be Partially Replaced in the Somatic mac.
To knock out the α-tubulin gene (ATU), we constructed pTUB-KO (Fig. 1) by replacing the whole coding region with the neomycin-resistance gene (neo) coding sequence, which confers pm resistance in T. thermophila (22). In this plasmid, the neo coding region is flanked by the 5′ and 3′ ATU gene flanking sequences, which drive neo gene expression after transformation.
Since T. thermophila has only a single ATU gene (1), cells presumably would not be viable if all the ATU copies were knocked out in the transcriptionally active mac. To test the pTUB-KO construct and determine whether cells whose developing mac ATU genes had been partially disrupted could survive, a biolistic somatic transformation (14) was performed using linearized pTUB-KO. About 100 pm-r transformants were obtained that grew normally in up to 3 mg/ml and survived in up to 7 mg/ml pm. Wild-type cells were killed in 120 μg/ml pm. To examine if these transformants contained the knockout fragment, genomic PCR was performed for four randomly selected clones, KO1–4, with primers (5′TUBx and 3′TUBz) located in the ATU gene flanking sequences (Fig. 2 A and B). In addition to the 1.8-kb band amplified from the wild-type ATU gene, a 1.2-kb band, the size expected from the neo gene flanked by the ATU sequences, was detected in all four transformants but not in the wild type, indicating that the transformants contain the knockout construct. To determine if the wild-type ATU gene has been replaced by the disrupted gene through homologous recombination at the ATU locus, Southern blot analysis using a 5′-specific probe for the ATU gene was performed with HindIII- or HaeIII-digested genomic DNA isolated from the four transformants grown in 1 mg/ml pm (Fig. 2 A and C). In wild-type cells, the expected single band was detected with either HindIII (3.2 kb) or HaeIII (8.0 kb) digestion. In all transformants KO1–4, the intensity of the wild-type band was reduced and a smaller band, resulting from neo integrated into the ATU locus, was present. Comparing the intensities of the knockout and wild-type bands, more than 50% of the mac ATU genes were knocked out in each transformant, indicating that cells can survive with less than 50% of their mac ATU genes. These studies show that the pTUB-KO construct can be used to knock out the ATU gene and suggest macs derived from a mic containing one wild-type and one disrupted ATU gene should be viable.
Creation of a Germ-Line Knockout for the α-Tubulin Gene.
To create a biolistic germ-line transformant with a disrupted ATU gene (Fig. 3), conjugating B2086 and CU428 cells were bombarded with linearized pTUB-KO at 0.5-hr intervals from 3 to 4.5 hr after conjugation had been initiated, when germ-line transformation can occur (14). Bombarded cells were selected for pm resistance about 20 hr after bombardment, when conjugation is completed. Dozens of pm-r transformants were obtained for each time point and tested for resistance to mp. This drug selects for cells that have completed conjugation because CU428 is a heterokaryon containing the dominant Mpr gene only in the transcriptionally silent mic, which can be selected only after a successful mating, when it is expressed in the new mac and renders the cells resistant to mp (mp-r). One of the 4-hr pm-r transformants was also mp-r, indicating it was derived from a true exconjugant. Since cells that have completed conjugation are immature and cannot mate again for about 40–60 fissions (16), vegetative progeny of this G1 conjugant were single cell subcloned and each subclone was grown without drug selection for about 60 generations before being analyzed genetically. Note that macs and mics of clones derived from each of the four cells (karyonides) that develop from a conjugant pair have the same genotype but each represents an independent line of mac development (16). Since the cells were plated before pairs had separated into exconjugants, progeny of all four karyonidal clones should be found in the same microtiter well. Although they are genetically identical, because mating type is determined epigenetically, the progeny of these karyonidal clones can have different mating types.
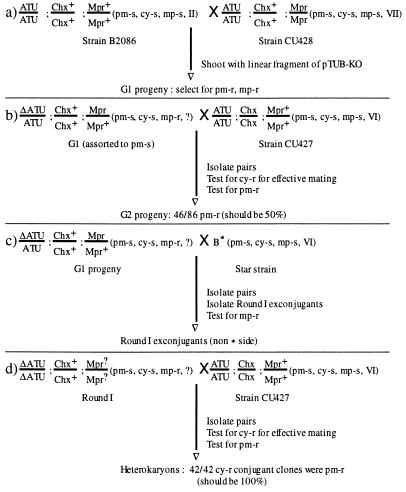
Figure 3.
Creation and testing of knockout heterokaryons homozygous for disrupted ATU genes in the mic (ΔATU) and containing only wild-type ATU genes in the mac. Chx is a dominant gene conferring resistance to cycloheximide (cy-r), whereas the wild-type Chx+ is sensitive (cy-s). Mpr is a dominant gene conferring resistance to 6-methylpurine (mp-r), to which cells with the wild-type Mpr+ gene are sensitive (mp-s). Roman numerals indicate mating types; mating types not determined are indicated by ?. Note that in cross d only a mutant ΔATU/ΔATU round I product of cross c is shown. An approximately equal number of nonmutant ATU/ATU round I cells are obtained that do not yield any pm-r progeny. The genotype of the Mpr locus in round I exconjugants was not determined. Values obtained for test crosses b and d are from a single subline.
To test whether the subclones were germ-line transformants—i.e., whether they can transfer the knockout marker to the next generation in a genetic cross—the G1 subclones were mated to CU427, a cy-s heterokaryon homozygous in the mic for the Chx gene conferring dominant cy-r. Most subclones gave progeny resistant to both cy and pm, demonstrating effective mating to CU427 and transfer of the knockout to the subsequent generation.
Genotypic Analysis of Phenotypically Assorted G1 Subclones.
The Tetrahymena mac is polyploid, with most genes present in ≈45 copies in G1 phase (23). During vegetative growth, the mac divides amitotically, distributing approximately equal numbers of the multiple copies of each gene randomly between the two daughter cells. This results in a phenomenon known as phenotypic assortment (24). After about 60 vegetative (i.e., asexual) divisions, most sublines derived from a heterozygous karyonidal clone have assorted; their macs contain exclusively one or the other of the two alleles (25). Once a subline expresses only one allele, it never reverts to the other type, though it can be demonstrated genetically that the mic is still heterozygous. Since the G1 cells obtained from the germ-line transformation had been grown for ≈60 generations in the absence of pm, most cells should have assorted the ATU genes, expressing either the wild-type gene or the knockout. Cells expressing the wild-type gene should be pm-s. Vegetative progeny of G1 subclones which gave pm-r and cy-r G2 progeny when mated to CU427 were single cell subcloned and the isolated sublines were grown in 120 μg/ml pm. About 30% of the sublines derived from each subclone failed to give any pm-r vegetative progeny, indicating that their macs contain only wild-type ATU genes. Three pm-s G1 sublines were mated again to CU427 to test for Mendelian inheritance (Fig. 3). About 90 conjugating pairs were isolated and grown up for each subline, and the G2 progeny derived from the pairs were selected first for cy-r and then for pm-r. In every cross, ≈50% of cy-r G2 were also pm-r (the fraction of cy-r G2 clones that were also pm-r in the three crosses was 46/86, 41/87, and 34/75), demonstrating that the knockout marker is inherited in a typical Mendelian fashion. Therefore, those G1 sublines are true germ-line transformants. The pm-s germ-line G1 sublines were mated to one another to test for different mating types. Two pm-s germ-line G1 clones of different mating types were obtained, named AKO2 and AKO5 for ATU knockout. Because they were derived from conjugating B2086 and CU428 cells, the latter of which are homozygous for Mpr, both clones were resistant to mp.
Construction of Heterokaryons Containing Homozygous Germ-Line Knockouts for the ATU Gene.
Star (*) strains have defective mics; they form conjugal pairs but cannot donate genetic material. When mated to a star strain, a normal strain donates a gametic nucleus but receives nothing in return when gametic nuclei are normally reciprocally exchanged. The single haploid nucleus in each conjugant then diploidizes and most cells separate without forming a new mac. These cells retain their old macronuclear phenotype and can be immediately remated but have new, homozygous micronuclear genotypes that depend on the genetic composition of the normal parent and which meiotic product was (randomly) selected to form gametic nuclei. This process is referred to as round 1 of genomic exclusion (16) (Fig. 3).
Both AKO2 and AKO5 could form pairs with the strain B* (VI) which is mp sensitive. Single pairs were isolated and exconjugants were each isolated into individual drops of medium and treated with mp to select for the “nonstar” or normal parent side (Fig. 3). Two clones, one derived from AKO2 and one from AKO5, were obtained which, when crossed to CU427, gave cy-r progeny all of which were also pm-r, indicating that both copies of the ATU gene had been knocked out in the diploid mics of these two clones. Since they are pm-s, that is, their macs contain only the wild-type ATU gene, the two round 1 clones (AAKO2 and AAKO5) are the desired heterokaryons.
Test of Gene Transfer from Somatic to Germ-Line (and Among Somatic) Nuclei in T. thermophila.
During conjugation, new macs in the karyonides develop from the fertilized mics in the conjugants, and all the genes in the new macs are inherited from the fertilized mics. However, it has never been tested whether DNA can be transferred from somatic macs to germ-line mics in vegetative cells or from the old mac back to the mic prior to completion of the second postzygotic division or to the developing macs during conjugation. Since transformation of mics and of new macs with exogenous DNA during conjugation is easily accomplished (8, 14), it is possible that DNA lost from the degenerating old mac gets into the developing mics during the early stages of conjugation, or into the developing macs during macronuclear development.
To test for gene transfer, two mating types of a heterokaryon homozygous for lethal (or recessive) alleles in the mic and a viable (or dominant) allele in the mac are required. When such cells mate, only a gene transfer from the mac would enable them to produce viable (or dominant) progeny. The creation of heterokaryons homozygous for knockouts of the essential ATU gene allows this test. Because both copies of the gene are knocked out in the diploid mics of the heterokaryons AAKO2 and AAKO5, when they mate the developing new macs do not receive a copy of the ATU gene, and the progeny lacking α-tubulin proteins should die. Only if ATU genes from the wild-type macs of the heterokaryons are transferred to mics or to the developing macs can the progeny obtain an ATU gene and survive.
To perform this gene transfer test (Table 1), 1 × 107 cells each from strains AAKO2 and AAKO5 were mated. Twenty-one hours after mixing, 1 × 107 exconjugants were obtained that contained two new or two new and one old macs, and cells were refed. Four hours later, pm (120 μg/ml) was added. The two heterokaryons were also mated to the wild-type strain B2086 to test mating ability, and a cross between wild-type strains B2086 and CU428 was performed as a negative control. No pm-r exconjugants were isolated from 1.0 × 107 conjugants obtained from the cross AAKO2 × AAKO5. However, 5.2 × 106 conjugants from the cross AAKO2 × B2086, and 6.3 × 106 conjugants from the cross AAKO5 × B2086, were pm-r, indicating that the heterokaryons are normal and competent in conjugation. As expected, none of the conjugants from the wild-type cross B2086 × CU428 was pm-r. This test was repeated once with similar results. Thus, in about 2 × 107 exconjugants in two tests, which contain about 4 × 107 developing macs, gene transfer from somatic macs to germ-line mics or from the old mac to the new mac was not observed. If such transfers occur at the ATU locus at all, they are very rare. These results also demonstrate again that the ATU gene in T. thermophila is essential, since progeny of knockout heterokaryons whose ATU genes were all disrupted could not survive.
Table 1.
Summary of gene transfer tests
| Cross | Total no. of cells | Mating efficiency, % | Total no. of conjugants | Total no. of pm-r cells |
|---|---|---|---|---|
| Experiment 1 | ||||
| AAKO5 × AAKO2 | 2.0 × 107 | 52 | 1.0 × 107 | 0 |
| B2086 × CU428 | 1.0 × 107 | 76 | 0.76 × 107 | 0 |
| B2086 × AAKO5 | 1.0 × 107 | 73 | 0.73 × 107 | 0.63 × 107 |
| B2086 × AAKO2 | 1.0 × 107 | 58 | 0.58 × 107 | 0.52 × 107 |
| Experiment 2 | ||||
| AAKO5 × AAKO2 | 2.0 × 107 | 57 | 1.1 × 107 | 0 |
| B2086 × CU428 | 1.0 × 107 | 72 | 0.72 × 107 | 0 |
| B2086 × AAKO5 | 1.0 × 107 | 70 | 0.70 × 107 | 0.59 × 107 |
| B2086 × AAKO2 | 1.0 × 107 | 61 | 0.61 × 107 | 0.36 × 107 |
Transformation of Conjugating Heterokaryons with an ATU Gene.
Our main aim in making these heterokaryons is to study the function of primary sequence elements of α-tubulin by transforming conjugating heterokaryons with ATU genes containing mutations of interest and examining the phenotypes of transformants. Because progeny of such a conjugation lack an intact essential ATU gene, they do not grow. However, they should be able to grow if they are transformed during conjugation with an ATU gene that supports growth. The only ATU gene expressed in these transformants should be the one introduced by transformation, since there is no transfer of the wild-type gene present in the old mac to the mic or the developing mac. Because cells that complete conjugation receive only disrupted ATU genes expressing the neo gene and the parental AAKO2 and AAKO5 cells are pm-s, pm can be used to select for progeny of successful conjugation.
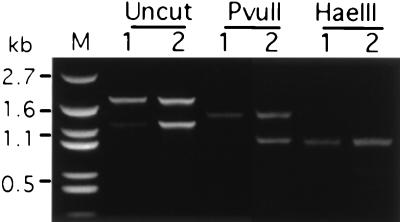
To test these expectations, conjugating AAKO2 and AAKO5 cells were biolistically transformed 11 hr after mixing (mac development II stage) (14, 19, 26) with linearized pTUB100E3-PvuII containing a wild-type ATU gene silently marked by HaeIII and PvuII sites (Fig. 2A). Two transformants survived in pm (120 μg/ml), suggesting their new macs received both the knockout gene, which rendered them pm-r, and an ATU gene, which enabled them to grow. To determine whether all ATU copies in the transformants were the exogenous gene introduced during transformation, the ATU genes in the two clones were amplified using genomic PCR (Fig. 4). Two fragments were amplified whose sizes correspond to the wild-type ATU gene (1.8 kb) and the disrupted gene (1.2 kb). When digested with either HaeIII or PvuII, the 1.8-kb band was cleaved to smaller size fragments and the digestion was complete. Therefore, the only ATU gene in the mac of the two transformants is the one introduced by transformation. The absence of wild-type ATU genes in these cells demonstrates that the germ-line transformants were true knockouts created by homologous replacement of the wild-type ATU gene by the disrupted gene and that the germ-line mics of AAKO2 and AAKO5 were homozygous for the disrupted ATU gene. Thus our strategy of eliminating the wild-type α-tubulin genetic background through transformation of conjugating knockout heterokaryons was successful. The transformants are stable as expected and have been in culture for hundreds of generations since they were created. We also have obtained viable pm-s subclones from these transformants, suggesting that the wild-type ATU gene replaced some of the disrupted genes, allowing phenotypic assortment.
Figure 4.
Genomic PCR analyses of transformants obtained from conjugating ATU knockout heterokaryons transformed with a wild-type ATU gene. Conjugating knockout heterokaryons were transformed with linearized pTUB100E3-PvuII (see Fig. 2A). Genomic PCR was performed with primers 5′TUBx and 3′TUBz (shown in Fig. 2A) for two transformants (1 and 2) which survived in pm. The 1.8-kb band of the undigested PCR product could be either the endogenous or the transforming ATU. The 1.2-kb band corresponds to the knockout fragment containing the neo gene. The 1.8-kb band in both transformants completely shifts to small sizes after digestion by either enzyme, indicating that the cells contain only the transforming ATU gene. The 1.2-kb band was also cut by the two enzymes into small fragments as expected. Lane M, PUC19 marker (Biosynthesis, Lewisville, TX).
We tested several parameters for the rescue of heterokaryon conjugants with a wild-type ATU gene. The timing of transformation appears to be critical. When we transformed unfed exconjugants from the cross between the two heterokaryons at about 24 hr after mixing, the rescue efficiency was greatly increased to as high as about 300 transformants per μg of DNA. Using more exconjugants also resulted in more transformants. We obtained 900 transformants per μg of DNA with 3 × 107 mated heterokaryon cells, compared with about 300 transformants per μg of DNA with 1 × 107 cells from the same cell population. Increasing the amount of DNA yielded more transformants without significantly affecting the number obtained per μg of DNA using 2, 4, or 8 μg of DNA. Thus, knockout heterokaryons can be rescued in large numbers at high efficiency with an ATU gene after optimization for transformation. This rescue efficiency should enable us to analyze mutant ATU genes and even to mutagenize the ATU coding region randomly in vitro, transform knockout heterokaryon exconjugants, and screen for mutations that affect microtubule assembly and functions.
DISCUSSION
Because macs are polyploid and homologous recombination events are frequent in Tetrahymena, we found that when they are transformed with highly disadvantageous mutations associated with flanking selectable markers, the cells routinely execute a crossover with the endogenous gene between the selectable marker and the deleterious mutation. This creates a gene with the selectable marker and the wild-type sequence at the site of the deleterious mutation whose function was being analyzed, defeating the purpose of the experiment. Crossing-out of deleterious mutations was observed for essential genes (e.g., the Tetrahymena α-tubulin gene), and for redundant genes [e.g., the Tetrahymena β-tubulin genes for which there are two copies (L. Gu and M.A.G., unpublished results)], suggesting this is a common problem for mutagenic analysis using macronuclear transformation. In this paper, we have described a novel strategy for introducing genes into macs lacking the gene being studied, thereby eliminating the possibility of homologous recombination between the introduced and endogenous genes. We created knockout heterokaryons having both copies of an essential gene knocked out in the mic while retaining wild-type genes in the mac. These cells must accept an introduced gene (as long as it is viable) or die and should allow the phenotypic effects of deleterious mutations to be examined. This strategy can also be extended to small multigene families, provided all of the genes in the family can be knocked out in the mic. This would require germ-line transformation to knock out the genes individually and then mating different knockouts to obtain a strain containing germ-line knockouts for all of the genes that are functionally redundant.
Knockout heterokaryons make it easier and faster than previous transformation methods to obtain transformants containing only the mutant version of the gene being analyzed. Transformation by microinjection (27) or conjugant electroporation (10) results in transformation of only some macronuclear copies of the gene. To eliminate all endogenous wild-type copies, prolonged growth under selection pressure is required. Furthermore, to prove a complete replacement in which the mutation is totally responsible for the phenotype to be observed, usually Southern and Northern blot analyses have to be performed, which are again time consuming. However, transformation of knockout heterokaryon conjugants results in viable progeny that have the transforming gene as the only functional form of the genes, because the endogenous mac copies are all disrupted, obviating the need for a selectable marker in the transforming DNA. Thus, as soon as a transformant is obtained, it can be used for phenotypic analysis. Also the fact that complete replacements are selected immediately rather than after a period of vegetative growth should greatly decrease the chance of suppressor mutations occurring to confuse the analysis.
It is not known whether gene transfer from soma to germ line occurs in nature. A rigorous test for this is difficult because multicellular organisms are hard to mate in large numbers and, in unicellular organisms such as yeast, the somatic and the germ-line nuclei develop sequentially and are never contemporaneous. In ciliated protozoa, the co-existence of germ-line and somatic nuclei in the same cytoplasm coupled with new molecular tools for germ-line and somatic transformation offer a unique opportunity for a test of gene transfer from soma to germ line. Interactions between the somatic and germ-line genomes in Tetrahymena have recently been demonstrated by Chalker and Yao (28), who showed that when the somatic nucleus of T. thermophila was loaded with a specific germ-line-limited sequence by microinjection, site-specific DNA deletion of that sequence during subsequent mac development was specifically blocked. This failure to excise mic-specific elements was inheritable in a non-Mendelian fashion. These results strongly suggest that DNA of the somatic mac can affect DNA rearrangement in the germ-line mic during conjugation. Similar phenomena have also been observed in another ciliate, Paramecium (see ref. 13 for review). One possible explanation of these results is that the mic-limited DNA elements escape from old macs and diffuse into the cytoplasm or into the developing mac to compete for limiting trans-acting factors. If that is the case, it might be possible for the escaped somatic mac DNA to reenter the germ line and to be transmitted to the new macs, as exogenous DNA does during somatic and germ-line transformation. The creation of heterokaryons which are homozygous for germ-line knockouts of the essential α-tubulin gene allowed a test of gene transfer from somatic to germ-line nuclei in Tetrahymena. Gene transfer from somatic macs to germ-line mics or from old macs to new macs at the ATU locus was not observed in about 2 × 107 exconjugants in two tests. There are three possible explanations for this result: (i) DNA is not transferred; (ii) gene transfer occurs at other loci but not at the ATU locus; and (iii) gene transfer occurs at the ATU locus but is very rare and the number of exconjugants we have tested was not large enough to detect these events. Since the mic ATU locus can be transformed by exogenous DNA, it seems unlikely that DNA escapes frequently from the mac to transform the mic during vegetative growth or conjugation. Additional studies using knockout heterokaryons to study the ATU and other essential genes should determine whether transfer from somatic to germ-line nuclei occurs at other loci or with detectable frequency at the ATU locus.
Acknowledgments
We are grateful to Dr. Peter J. Bruns and Donna Cassidy-Hanley for consultation and discussion. This work was supported by Public Health Service Grant GM26973 from the National Institutes of Health.
Footnotes
Abbreviations: pm, paromomycin; cy, cycloheximide; mp, 6-methylpurine; cy-r, cy-resistant; pm-r, pm-resistant; pm-s, pm-sensitive; mac, macronucleus; mic, micronucleus.
References
- 1.Gaertig J, Thatcher T H, McGrath K E, Callahan R C, Gorovsky M A. Cell Motil Cytoskeleton. 1993;25:243–253. doi: 10.1002/cm.970250305. [DOI] [PubMed] [Google Scholar]
- 2.Bré M-H, de Nechaud B, Wolff A, Fleury A. Cell Motil Cytoskeleton. 1994;27:337–349. doi: 10.1002/cm.970270406. [DOI] [PubMed] [Google Scholar]
- 3.Penque D, Galego L, Rodrigues-Pousada C. Eur J Biochem. 1991;195:487–494. doi: 10.1111/j.1432-1033.1991.tb15729.x. [DOI] [PubMed] [Google Scholar]
- 4.Bulinski J C, Gunderson G G. BioEssays. 1991;13:285–293. doi: 10.1002/bies.950130605. [DOI] [PubMed] [Google Scholar]
- 5.Piperno G, Ledizet M, Chang X J. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redeker V, Levilliers N, Schmitter J-M, Le Caer J-P, Rossier J, Adoutte A, Bré M-H. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- 7.Ludueña R F, Banerjee A, Khan I A. Curr Opin Cell Biol. 1992;4:53–57. doi: 10.1016/0955-0674(92)90058-k. [DOI] [PubMed] [Google Scholar]
- 8.Gaertig J, Gorovsky M A. Proc Natl Acad Sci USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaertig J, Thatcher T H, Gu L, Gorovsky M A. Proc Natl Acad Sci USA. 1994;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaertig J, Gu L, Hai B, Gorovsky M A. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaertig J, Cruz M A, Bowen J, Gu L, Pennock D G, Gorovsky M A. J Cell Biol. 1995;129:1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovlie A, Haller B L, Orias E. Proc Natl Acad Sci USA. 1988;85:5156–5160. doi: 10.1073/pnas.85.14.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer E, Duharcourt S. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- 14.Cassidy-Hanley, D., Bowen, J., Lee, J., Cole, E. S., VerPlank, L. A., Gaertig, J., Gorovsky, M. A. & Bruns, P. J. (1997) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 15.Allen S L. Science. 1967;155:575–577. doi: 10.1126/science.155.3762.575. [DOI] [PubMed] [Google Scholar]
- 16.Bruns P. In: The Molecular Biology of Ciliated Protozoa. Gall J G, editor. Orlando, FL: Academic; 1986. pp. 27–44. [Google Scholar]
- 17.Gorovsky M A, Yao M-C, Keevert J B, Pleger G L. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 18.McGrath K E, Yu S-M, Heruth D P, Kelly A A, Gorovsky M A. Cell Motil Cytoskeleton. 1994;27:272–283. doi: 10.1002/cm.970270308. [DOI] [PubMed] [Google Scholar]
- 19.Johnson K A. Methods Enzymol. 1986;134:306–317. doi: 10.1016/0076-6879(86)34098-9. [DOI] [PubMed] [Google Scholar]
- 20.Martindale D W, Allis C D, Bruns P J. Exp Cell Res. 1982;140:227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- 21.Orias E, Bruns P J. In: Methods in Cell Biology. Prescott D M, editor. New York: Academic; 1975. pp. 247–282. [Google Scholar]
- 22.Kahn R W, Andersen B H, Brunk C F. Proc Natl Acad Sci USA. 1993;90:9295–9299. doi: 10.1073/pnas.90.20.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorovsky M A. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- 24.Nanney D L. Experimental Ciliatology. New York: Wiley; 1980. [Google Scholar]
- 25.Doerder F P, Deak J C, Lief J H. Dev Genet. 1992;13:126–132. doi: 10.1002/dvg.1020130206. [DOI] [PubMed] [Google Scholar]
- 26.Orias E. In: The Molecular Biology of Ciliated Protozoa. Gall J G, editor. Orlando, FL: Academic; 1986. pp. 45–84. [Google Scholar]
- 27.Tondravi M M, Yao M C. Proc Natl Acad Sci USA. 1986;83:4369–4373. doi: 10.1073/pnas.83.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalker D L, Yao M C. Mol Cell Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]