Abstract
The status of the Archaea as one of the three primary Domains emphasizes the importance of understanding their molecular fundamentals. Basic transcription in the Archaea resembles eucaryal transcription. However, little is known about transcriptional regulation. We have taken an in vivo approach, using genetics to address transcriptional regulation in the methanogenic Archaeon Methanococcus maripaludis. We identified a repressor binding site that regulates nif (nitrogen fixation) gene expression. The repressor binding site was palindromic (an inverted repeat) and was located just after the transcription start site of nifH. Mutations that changed the sequence of the palindrome resulted in marked decreases in repression by ammonia, even when the palindromic nature of the site was retained. The same mutations greatly decreased binding to the site by components of cell extract. These results provide the first partial description of a transcriptional regulatory mechanism in the methanogenic Archaea. This work also illustrates the utility of genetic approaches in Methanococcus that have not been widely used in the methanogens: directed mutagenesis and reporter gene fusions with lacZ.
Keywords: Methanococcus, nif genes
The realization that living organisms can be divided into three main lineages, the domains Bacteria, Archaea, and Eucarya (1), led to a quest to discover the molecular fundamentals that distinguish them. Archaea, constituting one of the two prokaryotic domains, have a transcriptional apparatus that seems to reflect their phylogenetic relatedness to Eucarya. Thus, their RNA polymerases resemble the eucaryal RNA polymerases in subunit complexity, sequence and immunological similarity, promoter type recognized, and association with general initiation factors (2). Homologues of the TATA box-binding protein and the transcription factor TFIIB have been identified in Archaea (3–7). Furthermore, human and yeast TATA box-binding proteins replaced a required factor for in vitro transcription by RNA polymerase from Methanococcus thermolithotrophicus, a species closely related to the one used in this study (8).
Despite this emerging understanding of archaeal transcription, little is known about its mechanisms of regulation. Within a wide range of mechanisms, typically bacterial and typically eucaryal paradigms for transcriptional regulation can be distinguished (9–14), and one might expect yet another set of paradigms in the Archaea. For example, a typical eucaryal gene may be regulated by multiple activators that interact with various subunits of the initiation complex. Eucaryal repression can involve repressor binding throughout the promoter region and interference with various components of the transcription apparatus, including activators. In contrast, a typical bacterial gene may be regulated by a single activator that binds upstream from the promoter and/or by repressor binding at operator sites whose locations are restricted to the area near or downstream from the promoter. Bacterial repressors interfere directly with some step in transcriptional initiation. Unlike in Eucarya, bacterial repressors are typically dimers or tetramers, and operators are palindromic (inverted repeats).
To address the question of transcriptional regulation in the Archaea, we are using nitrogen fixation in Methanococcus maripaludis as a model system. M. maripaludis is a mesophilic, marine methanogen that is capable of diazotrophic growth (15, 16) and is one of a few archaeal species for which effective genetic methods are emerging. We report here that nif (nitrogen fixation) gene expression is regulated by repressor binding to a palindromic sequence situated just after the transcription start site of nifH.
MATERIALS AND METHODS
Growth of M. maripaludis.
Cultures were grown anaerobically at 30°C as described (16). Medium McC (17) was used for routine growth and maintenance of strains, while nitrogen-free medium (with N2 in the headspace, ref. 16) with or without added NH4Cl (to 10 mM) were used for preparation of cells on ammonia or N2, respectively. Puromycin (2.5 μg/ml) and l-arginine (2 mM) were added for growth of all mutant strains. Cells grown on ammonia were used 1 day after inoculation (OD600nm = 0.5–0.6), whereas cells grown on N2 were used 5–6 days after inoculation (OD600nm = 0.2–0.25).
Construction of Plasmids Containing Promoter–lacZ Fusions.
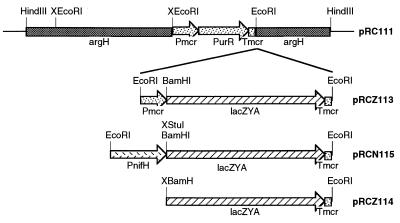
The EcoRI site of pGEM (Promega) was removed by digesting with EcoRI, filling in the ends with the Klenow fragment of DNA polymerase, and ligating to yield pGEM7.1. A 4.7-kb HindIII fragment containing the M. maripaludis argH gene was obtained from pKAS102 (18) and cloned into the HindIII site of pGEM7.1 to yield pRC100. The argH fragment served later as a site for homologous recombination into the M. maripaludis genome. One of two EcoRI sites within the 4.7-kb fragment (upstream from the argH gene) was removed from pRC100 by partial digestion with EcoRI followed by filling-in and ligating to yield pRC101. A 1.8-kb EcoRI fragment containing a puromycin resistance marker was obtained from pMudpur (16) and cloned into the remaining EcoRI site of pRC101 to yield pRC110. The EcoRI site upstream from the puromycin resistance fragment was removed from pRC110 as above to yield pRC111 (Fig. 1). The remaining EcoRI site of pRC111 was used to create three different constructs, all involving a promoterless lacZYA operon obtained from pSK202 (19) by digestion with SalI followed by filling-in, then digestion with BamHI followed again by filling-in. To create pRCZ113 (Fig. 1), the blunt-ended lacZYA fragment was cloned into the SmaI site of pMEB.1 (20) to yield pRCZ15, then an EcoRI fragment from pRCZ15 was cloned into pRC111. pRCZ113 (Fig. 1) contained lacZYA driven by the methylreductase promoter (Pmcr) from Methanococcus voltae and followed by the methylreductase terminator (Tmcr). To create pRCN115 (Fig. 1), the methylreductase promoter was removed from pRCZ15 by digesting with BamHI followed by filling-in, then digesting partially with EcoRI. Into this site was ligated a 1.2-kb fragment containing the M. maripaludis nifH promoter region that had been obtained from pMmp1 (16) by partial digestions with EcoRI and StuI. StuI cuts just downstream from the putative ribosome binding site of nifH. This generated pRCN35. The EcoRI fragment from pRCN35 was cloned into pRC111 to yield pRCN115. pRCN115 (Fig. 1) contained lacZYA driven by the M. maripaludis nifH promoter. To create pRCZ114 (Fig. 1), the promoterless lacZYA fragment was obtained from pRCZ15 by digestion with BamHI and EcoRI and ligated at one end to EcoRI-digested pRC111. The remaining ends were filled-in and ligated. pRCZ114 (Fig. 1) contained lacZYA without a promoter.
Figure 1.
Plasmids for introducing lacZ driven by various promoters into M. maripaludis. The X before a restriction enzyme indicates a site that was removed. Pmcr and Tmcr indicate the methylreductase promoter and terminator, PurR indicates the puromycin resistance gene, and PnifH indicates the nifH promoter.
Site-Directed Mutagenesis of the nifH Promoter Region.
The StuI–EcoRI fragment isolated from pMMP1 was cloned into pGEM7, where mutations were generated using the Transformer site-directed mutagenesis kit (CLONTECH). The primers used for the mutagenesis were 28–40 nucleotides long and contained the mutations shown in Fig. 2. The mutated promoter regions were obtained by digestion with EcoRI and BamHI and made into constructs for introduction into M. maripaludis as for pRCN115 above.
Figure 2.
Nucleotide sequence of nifH promoter region and mutations. The promoter and translational start sites are underlined, palindromic sequences overlined, and transcriptional start site indicated by Γ. Mutations are indicated in parentheses. Mutations altering the first, second, and both halves of the first palindrome are designated CT1, AG1, and CT1AG1, respectively. Similar mutations in the second palindrome are CT2, AG2, and CT2AG2. A mutation making the first palindrome identical to the second is TA3T, and a mutation deleting six nucleotides between the two palindromes is Δ6.
Transformation of M. maripaludis and Southern Blot Analysis of DNA Integration.
M. maripaludis transformation (21) was carried out with 5–10 μg of DNA. Similar results were obtained with supercoiled and linearized DNA, and supercoiled DNA was used routinely. Transformants were plated (18) with 2 mM arginine and 2.5 μg/ml puromycin. Individual colonies were streak-purified and inoculated into liquid medium. For Southern blot analysis, genomic DNA was prepared from 2 to 5 ml of culture. The cells were harvested by centrifugation and lysed by suspension in TE buffer. Proteinase K was added to 10 mg/ml, and the lysates were incubated at 50°C for 1 hr. Phenol–chloroform extractions were then performed, followed by ethanol precipitation. HindIII and EcoRI digests were probed with the 4.7-kb argH fragment and the lacZYA fragment, respectively. DNA transfer and hybridization were performed with nylon-based membrane (Zeta-probe, Bio-Rad) according to the manufacturer’s protocol. DNA fragments used as probes were isolated from agarose gels using Prep-A-Gene DNA purification kit (Bio-Rad) and labeled with Klenow enzyme using a random-primed DNA labeling kit (Boehringer Mannheim).
Primer Extension Analysis.
RNA was extracted from each culture by the guanidine–thiocyanate method followed by phenol–chloroform extraction and ethanol precipitation. RNA was extracted from 5 ml of culture for NH4+ grown cells, and from 20 ml of culture for N2 grown cells. Total RNA (50 μg) (determined spectrophotometrically) was used for each reaction. A 32P-labeled oligomer (0.2 μg) homologous to a portion of lacZ (5′-TAACGCCAGGGTTTTCCCAGT-3′) was used to prime cDNA synthesis by M-MLV reverse transcriptase (BRL). The products were run on a 6% acrylamide gel alongside a sequencing ladder.
Mobility Shift Assays.
DNA was PCR-amplified from pMmp1.1 (an EcoRI subclone of pMMP1, ref. 16) containing the wild-type palindromes or from a plasmid series analogous to pRCN115 containing the mutations. PCR primers were 5′-TCTAGAATTCTATACGCATAGTTCACC-3′ and 5′-GGAATTCTATATATTGTTGACTTTCGG-3′, except for mutations AG2 and CT2AG2, for which the second primer lacked the 3′ GG. Radioactive probe was produced by digesting the wild-type PCR product with EcoRI and filling-in with Klenow using 32P dATP. Extract from M. maripaludis grown on ammonia was prepared by lysing cells on ice with cold 50 mM Hepes (pH 7.5) and 5 mM DTT and removing debris by centrifugation at 4°C. Extract was kept in small aliquots at −70°C. Protein concentration was determined as in ref. 22. Cell extract (10 μg protein) was mixed with wild-type or mutant competitor DNA (2 μg), radiolabeled probe DNA (2 ng), and poly(dI·dC) (1 μg) in buffer (10 mM Hepes, pH 7.5/12% glycerol/10 mM DTT/300 μg/ml BSA), incubated at 30°C for 15 min, and run on a 4% acrylamide gel in Tris-glycine buffer (23).
RESULTS
Use of lacZ as a Reporter Gene to Monitor nifH Gene Expression.
To determine whether lacZ could be used as a reporter gene in M. maripaludis, we cloned a promoterless lacZYA operon (19) after the constitutive methylreductase promoter from Methanococcus voltae (20). This fusion construct, designated Pmcr-lacZ, was placed on a plasmid (Fig. 1, pRCZ113) containing a selectable marker for puromycin resistance (PurR, ref. 20). In this construct Pmcr-lacZ and PurR were flanked by two portions of a randomly cloned fragment of the M. maripaludis genome (18), later found to contain the argH gene (R.C.-K., unpublished work). This plasmid was introduced into M. maripaludis by transformation. Southern blot analysis showed that the plasmid had integrated into the genome by a single recombination event in the left portion of argH. When this strain was grown on agar medium containing 5-bromo-4-chloro-3-indolyl β-d-galactoside to detect β-galactosidase (β-gal) activity, blue color developed after the plates were exposed to air. Quantitative assay using o-nitrophenyl β-d-galactoside showed that the strain (Pmcr-lacZ, Table 1) had high β-gal activity as compared with wild-type M. maripaludis or a control strain containing the lacZYA operon without a promoter. The methylreductase promoter was then replaced with a 1.2-kb DNA fragment containing the nifH promoter region from M. maripaludis (Fig. 1, pRCN115). nifH encodes the nitrogenase reductase component of the nitrogenase complex, is transcribed from a typical archaeal promoter, and produces detectable mRNA only under nitrogen-fixing conditions (absence of NH4+, P. Kessler and J.A.L., unpublished work). The PnifH-lacZ construct was transformed into M. maripaludis, where β-gal assays showed marked repression by ammonia as expected (Table 1).
Table 1.
Effects of mutations in palindromes on nifH expression
| Construct or PnifH mutation | β-gal, ammonia-grown | β-gal, N2-grown | Ratio NH4+/N2 |
|---|---|---|---|
| Wild-type M. maripaludis | 2.4 ± 1.8 | 5.7 ± 3.5 | |
| Pmcr-lacZ | 282.0 ± 19.2 | 351.5 ± 106.8 | |
| No promoter-lacZ | 1.9 ± 0.8 | 2.0 ± 0.6 | |
| PnifH-lacZ | 3.1 ± 0.7 | 140.7 ± 17.9 | 0.01 |
| CT1 | 23.7 ± 6.6 | 34.7 ± 11.9 | 0.66 |
| AG1 | 62.3 ± 2.3 | 179.9 ± 57.0 | 0.34 |
| AG1 | 75.6 ± 3.4 | 290.1 ± 63.5 | 0.26 |
| CT1AG1 | 585.4 ± 41.9 | 844.4 ± 86.3 | 0.69 |
| TA3T | 5.2 ± 1.4 | 61.4 ± 28.3 | 0.05 |
| TA3T | 2.8 ± 0.7 | 172.7 ± 41.8 | 0.01 |
| CT2 | 13.1 ± 0.5 | 194.1 ± 34.1 | 0.06 |
| AG2 | 4.4 ± 1.9 | 96.8 ± 21.5 | 0.03 |
| CT2AG2 | 9.7 ± 1.7 | 356.8 ± 27.2 | 0.02 |
| Δ6 | 9.6 ± 1.0 | 393.0 ± 50.4 | 0.02 |
Each line presents data from a separate strain of M. maripaludis. Strains were grown in nitrogen-free medium under N2 in the presence (ammonia grown) or absence (N2 grown) of NH4+. Portions of cultures (0.2 ml) were used for β-gal measurements according to Miller (24). Values are averages from triplicate samples. Ratio NH4+/N2 was calculated after subtracting background. Additional experiments showed similar trends.
A series of M. maripaludis transformants analogous to that containing PnifH-lacZ was generated, each with a different mutation in the promoter region (see below). Southern blot analysis showed that in each transformant, integration of the construct had occurred in one of three configurations: integration of the entire plasmid into the left portion of argH, the same into the right portion of argH, or replacement of one copy of argH with the argH-flanked construct while simultaneously retaining a wild-type argH locus, perhaps on a different copy of the chromosome. In some cases several transformants were obtained with the same construct, and integration had occurred in different configurations. By comparing these, no consistent effect of configuration on β-gal activity could be discerned. The greatest difference observed was 3-fold (see two entries for TA3T, Table 1). Therefore, the PnifH-lacZ system could be used to measure marked changes in transcription from the nifH promoter.
A Specific Palindromic Sequence in the nifH Promoter Region Is Important for Repression.
Immediately following the start of transcription of nifH we found two sets of palindromic sequences reminiscent of bacterial repressor binding sites (Fig. 2). To test whether these sequences were involved in repression, we mutagenized specific nucleotides in and around the palindromes. We then generated M. maripaludis transformants containing these mutagenized promoter regions fused to the lacZ reporter gene as above. β-gal activities were measured after growth on NH4+ or N2 (Table 1). With NH4+, mutations that altered the first palindrome (CT1, AG1, and CT1AG1, see Fig. 2) resulted in clear derepression. Some mutations also altered β-gal activities during growth on N2 alone, and these effects were assumed to be due to changes in promoter strength, or in regulation by residual nitrogen or other factors. Therefore, to better evaluate the effects of the mutations with regard to ammonia repression, the results in the presence of ammonia were considered relative to N2 alone (ratio NH4+/N2, Table 1). Again, marked derepression occurred when the first palindrome was altered. Altering the second palindrome (CT2, AG2, or CT2AG2), making the first palindrome identical to the second (TA3T), or altering the spacing between the two palindromes (Δ6), had little or no effect. These results suggest that the first palindromic site, but not the second, is necessary for repression of nifH transcription by ammonia.
Palindromic regions have the potential to form secondary structures in the DNA or mRNA. However, our results show that the specific sequence of the palindrome, not merely the potential for secondary structure, is the important factor in nifH repression, since mutation AG1CT1 retained the palindromic nature of the site but caused marked derepression. The palindromic nature of the site, and the requirement for both halves of the palindrome for full repression, may therefore reflect the dimeric nature of a repressor protein that binds there.
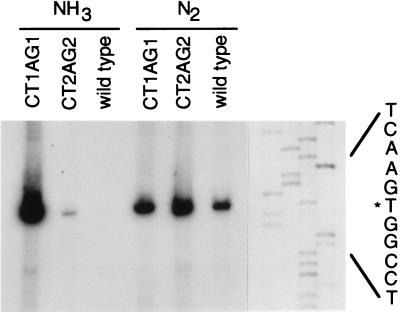
Assay of nifH-lacZ mRNA by primer extension analysis (Fig. 3) confirmed the results of β-gal measurements: only a mutation in the first palindrome caused marked derepression in the presence of ammonia. Primer extension analysis also showed that transcription initiated from the same site in all cases, eliminating the possibility that the mutations had introduced alternative promoters.
Figure 3.
Primer extension analysis of the nifH promoter–lacZ transcript produced from wild-type or mutant promoter regions during growth on ammonia or N2. Visualization of total RNA after gel electrophoresis indicated that the quantity of RNA was similar for all samples.
The Palindromic Sequence Is Necessary for Specific Binding of a Component in Cell Extract.
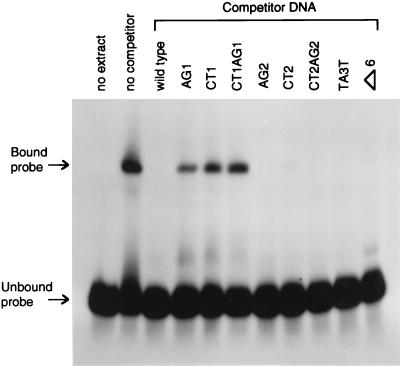
We used electrophoretic mobility-shift assays to show directly that a factor in M. maripaludis cell extract does indeed recognize and bind the first palindromic site. A fragment of the nifH promoter region spanning the two palindromic sites, but excluding the TATA box and the putative ribosome binding site, was radiolabeled and run on a gel (Fig. 4, no extract). Cell extract from ammonia-grown M. maripaludis caused a clear shift in the mobility of the DNA fragment (Fig. 4, no competitor). Unlabeled competitor DNA eliminated this mobility shift (wild-type competitor DNA). Different competitor DNA fragments containing the eight mutations were also used (see remaining lanes of Fig. 4). Competition was greatly decreased if and only if the competitor DNA contained the mutations altering the first palindrome. These results verified the role of the first palindromic site in the binding of a repressor.
Figure 4.
Electrophoretic mobility-shift assay for binding of M. maripaludis extract to wild-type and mutant palindrome-containing region. A 1000-fold excess of unlabeled competitor DNA was tested for its ability to prevent the mobility shift caused by binding of cell extract to the labeled wild-type region.
DISCUSSION
We implemented the use of a lacZ reporter gene to monitor nifH gene expression in M. maripaludis. [The use of the β-glucuronidase gene uidA as a reporter gene in M. voltae has also been reported (25), and we found that it worked in M. maripaludis as well]. Our results with wild-type and mutant nifH promoter regions demonstrated that nifH transcription is negatively regulated by ammonia, that a palindromic sequence immediately following the transcription start site is important for repression, and that a similar palindrome downstream from the first plays no major role. Primer extension analysis of PnifH-lacZ constructs confirmed the results from β-gal measurements. Furthermore, electrophoretic mobility-shift assays demonstrated specific binding of a component of cell extract to the first palindromic sequence.
The palindromic nature of the nifH repressor binding site in M. maripaludis, and its location immediately after the transcription start site, suggest a mechanism that is similar to certain classical paradigms. Many bacterial repressors bind as dimers or tetramers to palindromic operators, where the cooperative binding of two identical subunits to the two halves of the palindrome gives stability to the complex. In our system too, both halves of the repressor binding site were important, although some repression was apparently retained when only the second half of the palindrome was disrupted. Bacterial operators are typically positioned such that the bound repressor prevents RNA polymerase binding or interferes with some step in the initiation of transcription. For example, the lac operator contains a 19-bp palindromic sequence whose left end is positioned at the transcription start (26). By comparison, the palindromic sequence in the M. maripaludis nifH operator starts just two nucleotides from the transcription start, and repressor binding there could also interfere with some step in transcriptional initiation. We are aware of one other example of transcriptional regulation by repression in Archaea; a repressor present in phage φH lysogens of Halobacterium halobium apparently functions by binding to palindromic sites immediately upstream of the TATA box for a lytic gene (27, 28). However, despite similarities between bacterial repression and the observations made to date in Archaea, the picture is still incomplete and it would be premature to suggest that repression in Archaea occurs by a bacterial mechanism.
In some bacterial repression systems (e.g., lac and λ CI, refs. 29–31), and evidently in H. halobium φH as well (27), nearby copies of similar palindromes allow higher order cooperative interactions to occur due to repressor tetramers. In the M. maripaludis nifH promoter region, however, although a second palindrome exists, no evidence for any role was obtained. We did find a single copy of a similar palindrome in another location in M. maripaludis: the sequence GGAAAGCTATTTCC is centered about 21 bp downstream of a putative TATA element of glnA, another nitrogen-regulated gene (R.C.-K. and J.A.L., unpublished work). Binding of a repressor to a conserved palindromic sequence may be a central theme in nitrogen regulation in this Archaeon.
In M. maripaludis, regulation of nifH transcription by nitrogen presumably requires that repressor binding not occur in the absence of ammonia. Indeed, preliminary results from mobility shift assays similar to those reported here, but using extracts from N2-grown M. maripaludis, failed to show binding activity (C.B., unpublished work). Three possibilities remain to be tested: (i) repressor may not be present in cells grown on N2, (ii) an unidentified ligand prevents repressor binding under these conditions, or (iii) a ligand present in extracts of ammonia-grown cells facilitates binding.
The M. maripaludis nifH repressor must, of course, interfere with the archaeal transcriptional apparatus, which is similar to those of Eucarya. The general initiation factor TFIIB of eucaryal RNA polymerase II is now known to extend downstream from the TATA element in a complex that also contains the TATA box-binding protein (32). Homologous elements have been detected in Archaea, and if they assemble in the same configuration, the nifH repressor of M. maripaludis could interfere with the structure of this early complex.
Acknowledgments
We thank A. Klein for the plasmids pMEB.1 and Mip1. This work was supported by Grant 92-37305-7965 from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture.
Footnotes
References
- 1.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann P, Qureshi S A, Jackson S P. Trends Genet. 1995;11:279–283. doi: 10.1016/s0168-9525(00)89075-7. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi S A, Baumann P, Rowlands T, Khoo B, Jackson S P. Nucleic Acids Res. 1995;23:1775–1781. doi: 10.1093/nar/23.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowlands T, Baumann P, Jackson S P. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 5.Ouzounis C, Sander C. Cell. 1992;71:189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- 6.Creti R, Londei P, Cammarano P. Nucleic Acids Res. 1993;21:2942. doi: 10.1093/nar/21.12.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Wettach J, Gohl H P, Tschochner H, Thomm M. Proc Natl Acad Sci USA. 1995;92:472–476. doi: 10.1073/pnas.92.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 10.Buratowski S. Science. 1995;270:1773–1774. doi: 10.1126/science.270.5243.1773. [DOI] [PubMed] [Google Scholar]
- 11.Sauer F, Hansen S K, Tijan R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 12.Johnson A D. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 13.Cowell I G. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 14.Herschbach B M, Johnson A D. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 15.Whitman W B. In: Bergey’s Manual of Systematic Bacteriology. Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Vol. 3. Baltimore: Williams and Wilkins; 1989. pp. 2185–2190. [Google Scholar]
- 16.Blank C E, Kessler P S, Leigh J A. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitman W B, Shieh J, Sohn S, Caras D S, Premachandran U. Syst Appl Microbiol. 1986;7:235–240. [Google Scholar]
- 18.Sandbeck K A, Leigh J A. Appl Environ Microbiol. 1991;57:2762–2763. doi: 10.1128/aem.57.9.2762-2763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomich C-S C, Kaytes P S, Olsen M K, Patel H. Plasmid. 1988;20:167–170. doi: 10.1016/0147-619x(88)90022-4. [DOI] [PubMed] [Google Scholar]
- 20.Gernhardt P, Possot O, Foglino M, Sibold L, Klein A. Mol Gen Genet. 1990;221:273–279. doi: 10.1007/BF00261731. [DOI] [PubMed] [Google Scholar]
- 21.Tumbula D L, Makula R A, Whitman W B. FEMS Microbiol Lett. 1994;121:309–314. [Google Scholar]
- 22.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1983. [Google Scholar]
- 24.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 25.Beneke S, Bestgen H, Klein A. Mol Gen Genet. 1995;248:225–228. doi: 10.1007/BF02190804. [DOI] [PubMed] [Google Scholar]
- 26.Gralla J D. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 629–642. [Google Scholar]
- 27.Ken R, Hackett N R. J Bacteriol. 1991;173:955–960. doi: 10.1128/jb.173.3.955-960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolt P, Zillig W. Mol Gen Genet. 1992;235:197–204. doi: 10.1007/BF00279361. [DOI] [PubMed] [Google Scholar]
- 29.Oehler S, Eismann E R, Krämer H, Müller-Hill B. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis M, Chang G, Horton N C, Kercher M A, Pace H C, Scumacher M A, Brennan R G, Lu P. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 31.Ptashne, M. (1986, (1987) A Genetic Switch: Gene Control and Phage λ (Blackwell Scientific, Palo Alto and Cell Press, Cambridge, MA).
- 32.Nikilov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]