Summary
In the developing nervous system, the balance between proliferation and differentiation is critical to generate the appropriate numbers and types of neurons and glia. Notch signaling maintains the progenitor pool throughout this process. While many components of the Notch pathway have been identified, the downstream molecular events leading to neural differentiation are not well understood. We have taken advantage of a small molecule inhibitor, DAPT, to block Notch activity in retinal progenitor cells, and analyzed the resulting molecular and cellular changes over time. DAPT treatment causes a massive, coordinated differentiation of progenitors that produces cell types appropriate for their developmental stage. Transient exposure of retina to DAPT for specific time periods allowed us to define the period of Notch inactivation that is required for a permanent commitment to differentiate. Inactivation of Notch signaling revealed a cascade of proneural bHLH transcription factor gene expression that correlates with stages in progenitor cell differentiation. Microarray/QPCR analysis confirms the changes in Notch signaling components, and reveals new molecular targets for investigating neuronal differentiation. Thus, transient inactivation of Notch signaling synchronizes progenitor cell differentiation, and allows for a systematic analysis of key steps in this process.
Keywords: Notch activity, Hes1, Hes5, proneural bHLH transcription factor, progenitor cell, neural differentiation, insulinoma-associated 1
Introduction
The Notch signaling pathway is critical for many aspects of neural development. Notch-Delta signaling is thought to mediate most, if not all, lateral inhibitory interactions necessary for patterning neural cells (Lewis, 1996; Lowell, 2000). Notch activity in the retina is necessary in progenitor cells to maintain their undifferentiated state throughout the neurogenic period (Dorsky et al., 1995; Austin et al., 1995; Tomita et al., 1996; Henrique et al., 1997; Dorsky et al., 1997; Furukawa et al., 2000; Hojo et al., 2000; Satow et al., 2001; Silva et al., 2003; Takatsuka et al., 2004; Nelson et al., 2006; Jadhev et al., 2006; Yaron et al., 2006). Notch is also important in promoting the glial fate in multipotent progenitor cells, and may also play a role in the survival of neural stem and progenitor cells, and newly generated neurons (Gaiano and Fishell, 2002; Mason et al., 2006).
Despite the wealth of data on the functions of Notch signaling in development, there are some key aspects of this pathway that are not well understood. For example, while only a brief period of Notch signaling activation is required to cause multipotent neural crest stem cells to develop into glia (Morrison et al., 2000), no study has defined the period of time during which the Notch signal has to be inactive in order to cause neural differentiation. In addition, while many of the components of the Notch pathway have been identified in genetic screens, we know little of the cascade or kinetics of downstream molecular events that lead to neural differentiation following inactivation of this signaling pathway.
Analysis of the extensive number of mutant Notch alleles in Drosophila reveals that Notch signaling can be separated into two categories, canonical and non-canonical (reviewed by Martinez Arias, 2002). Canonical Notch signaling is active in lateral inhibition and depends upon DSL (Delta/Serrate (Jagged)/Lag) ligand-regulated binding of the extracellular domain of Notch (discussed by Chitnis, 2006). Binding of DSL ligands to Notch allows access of a presenilin/γ-secretase complex to cleave and release the Notch internal cytoplasmic domain (NICD). Then NICD translocates to the nucleus and forms a transcriptional activation complex with CSL/RBP-j□ and Mastermind (reviewed by Mumm and Kopan, 2000; Selkoe and Kopan, 2003). This activation complex positively regulates transcription of Notch target genes, such as the Hes genes, that act as effectors of Notch signaling.
The presenilin/γ-secretase complex necessary for canonical Notch signaling is composed of at least four proteins (presenilin, nicastrin, pen-2, and Aph-1) that regulate intramembrane proteolysis (RIPping) of type I membrane proteins (Chyung et al., 2005). All mammalian Notch family members (Notch1–4) require presenilin/γ-secretase-mediated release of their intracellular domains for their canonical activities (Saxena et al., 2001). Presenilin mutations are frequently used to analyze loss-of-function of the Notch signaling pathway (for example see Alexson et al., 2006; Mizuguchi et al., 2006). Additionally, γ-secretase inhibitors that have been developed largely as a means to treat Alzheimer's disease (reviewed by Tsai et al., 2002) have also been used to inhibit the Notch pathway. One γ-secretase inhibitor, DAPT, has been shown to phenocopy various Notch mutations in both zebrafish and Drosophila (Geling et al., 2002; Micchelli et al., 2003) and downregulates Hes1 and Hes5 gene expression and reporter activity (Ong et al., 2006; Nelson et al., 2006).
In this study, we have taken advantage of DAPT treatment to inactivate Notch signaling in retinal progenitors. We show that DAPT treatment causes a massive, synchronized differentiation of neural progenitors, leading to premature differentiation of stage-appropriate cell types. Temporal analysis of gene expression defines the cascade and kinetics of molecular changes that lead to neural differentiation. We define the amount of time that Notch must be inactivated that will lead to a permanent commitment of the progenitors to differentiate. We also show that a cascade of transiently and sequentially upregulated proneural bHLH transcription factor genes correlates with stages in neural differentiation. Microarray analysis confirms the early molecular changes in expression of Notch pathway components and identifies new immediate targets in the differentiation cascade. Thus, precise temporal control over neural progenitor cell differentiation allows systematic analysis of this process.
Methods
Animals and tissues
Fertilized white leghorn chicken eggs (Hyline) were incubated to embryonic day 4.5 (E4.5, stage 25; Hamilton and Hamburger, 1951) and pairs of eyes were collected in HBSS+ (Gibco/BRL). Extra-ocular tissues and pigmented epithelium were removed. Pairs of retinas were transferred to a 24-well plate (Falcon, 351147) and cultured (as described in Nelson et al., 2006) for 2–4 days at 37°C with nutation. Pairs of retinas were collected from embryonic day E12.5 and postnatal day P1 mice (C57Bl6 or Swiss Webster), and were cultured as above with gentle nutation. Tissue harvest was carried out according to approved protocols at the University of Washington. Mice were housed in the Department of Comparative Medicine. Insm1:LacZ mice are described in Breslin et al., 2003.
The γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT; Sigma) was used to inhibit the γ-secretase-dependent S3 cleavage of Notch, which releases the Notch internal cytoplasmic domain NICD (Geling et al., 2002). We previously demonstrated that DAPT induced neuronal differentiation in a concentration-dependent manner, with 10μM giving optimal results without precipitating in culture (Nelson et al., 2006). DAPT (10μM) was added to one retina, while an equal volume of DMSO was added to the sister retina as vehicle control. For some experiments, chick retinas were bisected and one half treated with DAPT, while the other half served as DMSO control. For transient inhibition of Notch signaling, E4.5 chick retinal explants were prepared as described above and incubated in the presence of DAPT (10μM) or DMSO for 1h, 3h, 6h, 12h, 24h, and 48h: explants were then washed with media three times at the respective timepoint and cultured for a total of 48h.
Quantification of changes in gene expression
Quantitative reverse transcriptase PCR (QPCR) was used to measure changes in gene expression levels due to DAPT treatment at 3h, 6h, 12h, 24h, and 48h of culture (as described in Nelson et al., 2006). Briefly, the lens and any remaining pigmented epithelium were removed, and total RNA was extracted with Trizol (Invitrogen) followed by digestion with RQ1 RNase-free DNase (Promega) and purified with RNeasy columns (Qiagen). This RNA served as template for oligo-dT-primed cDNA synthesis with SuperScriptII Reverse Transcriptase (RT; Invitrogen): an RT minus control reaction was also included for each sample. QPCR was performed with SYBR Green QPCR Master Mix (Applied Biosystems) and an Opitocon DNA Engine Real-Time QPCR machine (MJ Research). Sample concentrations were normalized to GAPDH according to the respective ratios of GAPDH levels per retinal pair, with three pairs of retinas analyzed per time point. Student's T-test was used to determine significance at each time point, ANOVA was used to determine significance between time points, and changes of P<0.05 were considered significant.
Microarray analysis was used to compare global gene expression changes between E14.5 mouse retinas treated with DAPT for 8h and DMSO controls. Total RNA was pooled from each condition (n=12 retinas per condition), and used to generate probes for hybridization to Affymetrix microarrays (Affymetrix Mouse Genome 430 2.0 Array; Center for Expression Arrays, University of Washington). QPCR was used to confirm changes of selected genes from the microarray. Total RNA isolated from three separate litters (E14.5–E15.5 mice, 9–12 retinas treated with DAPT or DMSO for 8h per condition, n=3), as prepared for the microarray study, was used for QPCR as described. The majority of mouse primers were obtained from PrimerBank (Wang and Seed, 2003; all primer sets are available upon request).
Transfections
E5.5 chick retinas were collected, dissociated by trypsin, triturated into single cells, and transfected with GFP control plasmid (Nelson et al., 2006) or NICD-IRES-GFP plasmid (Daudet and Lewis, 2005). Electroporation conditions were 25μg DNA per 400μl cells, 3 pulses, 537V, 50ms pulse length, 100ms pulse interval (2mm cuvettes, BTX T 820). Transfected cells were plated onto poly-D-lysine/laminin-coated coverslips and grown overnight in culture medium. DAPT (10μM) was added to one set of GFP- and NICD-transfected wells, while DMSO was added to the control set of wells. The cells were cultured for an additional 48h. After the culture period, cells were fixed with 4% paraformaldehyde for 30 min, immunolabeled with a rabbit anti-GFP antibody (1:2000, Dr. L. Berthiaume, University of Alberta), and goat-anti-rabbit ALEXA 488 (1:500, Molecular Probes).
Immunolabeling
Explants were fixed in 4% paraformaldehyde for 30 min at room temperature and immunolabeled as cryosections or wholemounts. Explants were prepared for cryosectioning by cryoprotecting through progressively higher sucrose concentrations before embedding in O.C.T. (Tissue-Tek). Sections and wholemounts were rinsed in PBS and blocked in 10% goat serum — 1× PBS-0.1% Triton X-100 (PBT). Primary antibodies include rabbit and rat anti-Phospho Histone 3 (PH3, 1:750 dilution, Upstate and Novus, respectively), rabbit anti-Visinin (1:3000, A. Polans, University of Wisconsin), rat anti-BrdU (1:200 dilution; Accurate Chemical), rabbit anti-Trß2 (1:500, D. Forrest, NIH), mouse anti-Pax6 (1:20 dilution) and mouse anti Isl1 (1:5 dilution; DHSB, Developmental Hybridoma Studies Bank), rabbit anti-Prox1 (1:1000 dilution, Chemicon), mouse anti-TUJ1 (1:750 dilution, Covance), mouse anti-Cyclin D3 (1:200 dilution, Lab Vision Corp.), rabbit anti-CRALBP (1:200 dilution, J. Saari, University of Washington), rabbit anti-Recoverin (1:1000 dilution, Chemicon), rabbit anti-Rho4D2 (rhodopsin, 1:1000 diltution, DHSB). For BrdU immunolabeling, sections were incubated with rat anti-BrdU antibody and DNase 1 (1:100, Sigma) overnight. Secondary antibodies were species-specific AlexaFluor 488 or 568 nm depending on the desired wavelength (1:500, Molecular Probes). Sections were mounted in Fluoromount-G (Southern Biotechnology Associates). Explants were mounted in 50% glycerol/PBS. Sections were imaged with an epifluorescent Zeiss Axioscope equipped with appropriate filter sets and Normarski/DIC optics and a Spot Camera, and/or a Zeiss Pascal laser scanning confocal microscope (LSCM, Carl M. Zeiss, Inc.). Explants were imaged with a fluorescent stereo dissecting scope (Nikon, eGFP/Texas Red filter set, equipped with a Spot Camera), and/or LSCM.
For activated Notch1 (actN1) immunolabeling, a modified protocol based on that described in Tokunaga et al., (2004) was used. Briefly, 6μn paraffin sections from E14.5 mouse embryo that received a 1h pulse of BrdU in utero prior to sacrifice were deparaffinized and rehydrated. Antigen retrieval was accomplished by autoclave treatment (5 min, 105°C) in TE buffer (10mM TrisCl, 1mM EDTA, pH9.0). Sections were washed with PBS, blocked in 10% goat serum in PBT for 1h, incubated with rabbit-actN1antibody (1:500, cleaved Notch 1 Val1744, Cell Signaling Tech.) overnight, washed 4× with PBS, incubated with goat-anti rabbit:alkaline phosphatase (1:500, Sigma) for 1h, washed 4× with PBT, equilibrated with NTMT, pH9.0, and incubated in NBT/BCIP substrate (Sigma). Sections were washed in PBS and subjected to sequential immunolabeling and fluorescent detection with primary and secondary antibodies as described, followed by Dapi counterstaining and mounting.
Results
Kinetics of Notch signaling inactivation
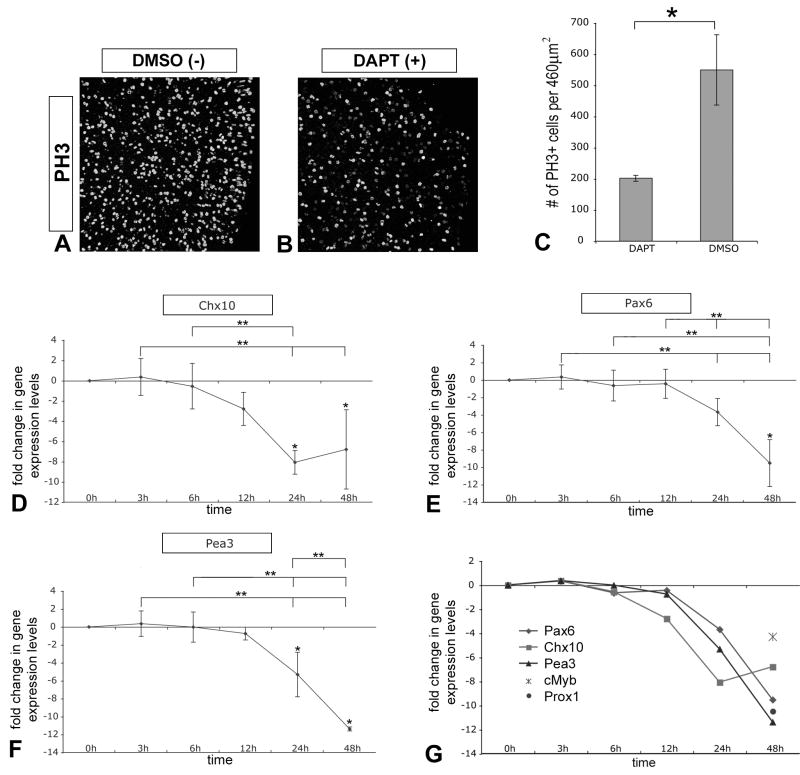
To determine the time-course of molecular changes due to loss of Notch activity, we treated embryonic day 4.5 (E4.5) chick retinal explants with DAPT. Pairs of retinal explants were cultured for 3 hours (3h), 6h, 12h, 24h, and 48h; one retina received DAPT (10□ M), while the sister retina served as the DSMO vehicle control (Fig 1 A). Gross morphological observations indicate that at 24h, the retina treated with DAPT was slightly smaller in size compared to its sister control, and appeared more compacted and ruffled. We quantified the levels of Notch-regulated genes by quantitative RT-PCR (QPCR). Data is presented as the average fold change between the DAPT-treated retina and control retina, normalized to GAPDH levels (Fig 1 B–F). The inactivation of Notch signaling caused a dramatic and rapid downregulation of Hes5 expression (Fig 1 B). This decline in Hes5 expression occurred as early as 3h, and was maintained throughout the culture period. There was also a 2-to 3-fold decrease in Hes1 expression in DAPT-treated retinas from 12h to 48h (Fig 1C). DAPT had relatively little effect on Notch1 expression itself, although a decrease was apparent by 48h (Fig 1D). Expression levels of Myt1, a Notch antagonist (Mueller et al., 1995; Bellefroid et al., 1996; Matsushita et al., 2002; Price et al., 2002), showed a transient ∼4-fold upregulation from 12 to 24h (Fig 1E). Comparing the relative changes in expression levels within this set of genes reveals an intriguing pattern (Fig 1 F). Inactivation of Notch signaling leads to a rapid reduction in the positive effectors of this pathway (Hes5 and Hes1), and a later transient increase in an antagonist of this pathway (Myt1), all of which would act to promote neural differentiation.
Figure 1. Kinetics of Notch signaling inactivation.
(A) Embryonic day E4.5 chick retinal explant pairs were collected and one explant cultured in the presence of DAPT (10□M), while the sister explant was cultured in DMSO as a vehicle control. Images are of typical eye pairs and were taken at the indicated times. By 24h of culture, the explant treated with DAPT began to appear smaller and exhibit aberrant morphology, both of which became more apparent by 48h. (B–E) QPCR was used to analyze changes in Hes5 (B), Hes1 (C), Notch1 (D), and Myt1 (E) gene expression levels over time due to inhibition of Notch signaling. For all QPCR experiments, three pairs of explants were analyzed per timepoint, error bars represent standard deviation from the mean, single asterisk indicates a significant difference between DAPT-treated explant compared to control at each timepoint as determined by one-way student's T-test, double asterisks indicate significant changes between timepoints as determined by ANOVA, and differences were considered significant at P<0.05. (F) Relative comparison of changes in gene expression levels over time. Note that Hes5 expression levels are dramatically reduced by 3h, and that Myt1 expression levels are transiently increased between 6–12h after Notch signaling inactivation.
Loss of Notch signaling reduces proliferation and progenitor gene expression
To further characterize the effects of the loss of Notch activity, we analyzed DAPT-treated E4.5 chick retinal explants for changes in proliferation and progenitor gene expression. Control and DAPT-treated retinas were labeled as wholemounts for the mitotic marker phospho-Histone 3 (PH3) and analyzed by laser scanning confocal microscopy (LSCM). Retinas treated with DAPT for 48h showed a large reduction in PH3+ progenitor cells compared to sister control retinas treated with vehicle alone (Fig 2 A, B). Quantification of this effect revealed ∼3-fold inhibition of proliferation due to loss of Notch activity (Fig 2 C). To ensure that DAPT was not cytotoxic to progenitor cells, we analyzed cell death after 6h of culture and found no significant difference in the number of propidium iodide (PI) labeled cells between DAPT and DMSO treated explants (Sup Fig 1 A–C).
Figure 2. Loss of Notch signaling reduces proliferation and progenitor gene expression.
(A, B) E4.5 chick retinal explants pairs cultured for 48h in DAPT or DMSO were wholemount immunolabeled with anti-phospho Histone 3 (PH3) antibody to reveal mitotic progenitor cells at the apical surface of the retina: images were acquired from flatmounted explants with LSCM. (C) Quantification of PH3+ progenitor cells indicated that DAPT treatment significantly reduced proliferation ∼3-fold compared to control; error bars indicate standard error of the mean, n=3 pairs, P<0.0186. (D–F) QPCR was used to analyze changes in Chx10 (D), Pax6 (E), and Pea3 (F) gene expression levels over time due to DAPT treatment as described before. (G) Relative comparison of changes in progenitor gene expression levels over time, including Prox1 and cMyb levels at the 48h timepoint. Note that decline in expression levels are apparent by 24h of Notch signaling inactivation.
We also analyzed levels of progenitor gene expression by QPCR as described above. Chx10, Pax6, Pea3, c-Myb, and Prox1 are all genes expressed in retinal progenitor cells (Green et al., 2003; Marquardt et al., 2001; Dyer et al., 2003; McCabe et al., 2006; Nelson and Reh, unpublished observations). Analysis of Chx10, Pax6, and Pea3 expression levels over time indicates that between 12h and 24h progenitor cell gene expression begins to decline; by 48h expression levels of all five progenitor genes had significantly decreased (Fig 2 D–G). Thus inhibition of Notch signaling leads to a decrease in progenitor cell gene expression and reduction in proliferation.
Loss of Notch signaling synchronizes neuronal differentiation
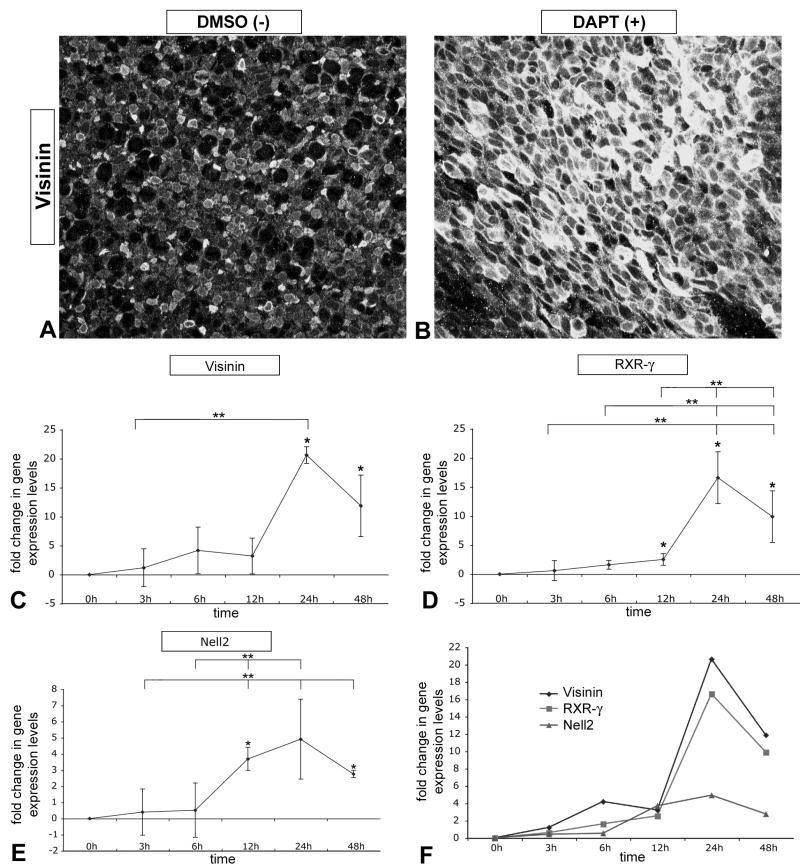
The loss of Notch activity causes a reduction of progenitor cells, and therefore should lead to an increase in neural differentiation. In the vertebrate retina, the first cell type to differentiate is the ganglion cell (see Hartenstein and Reh, 2001 for review). We previously observed that loss of Notch activity at embryonic day 3 (E3) caused an increase in ganglion cell differentiation (Nelson et al, 2006). To assess the timing of neural differentiation in E4.5 DAPT-treated explants, we measured gene expression levels of Nell2 by QPCR. Nell2 is a gene upregulated early during neural differentiation (Nelson et al 2002; 2004). Similar to Myt1, expression of Nell2 is significantly upregulated between 6h and 12h, and it maintains elevated expression levels throughout the duration of the culture (Fig 3 E).
Figure 3. Loss of Notch signaling synchronizes neuronal differentiation.
(A, B) E4.5 chick retinal explants pairs cultured for 48h in DAPT or DMSO were wholemount immunolabeled with anti-Visinin antibody to reveal differentiating cone photoreceptors in the apical retina: images were acquired from flatmounted explants with LSCM. (C–E) QPCR was used to quantify changes in gene expression levels of Visinin (C) and RXR-□ (D), both of which are specifically expressed in differentiating cones in chick, and Nell2 (E), which is generally expressed during neuronal differentiation, including ganglion cells and photoreceptors (Nelson et al., 2002; Nelson and Reh, unpublished observations). (F) Relative comparison of changes in neuronal gene expression over time as described before. Note that the general neuronal differentiation gene is upregulated by 12h after Notch signaling inactivation, while cone specific genes are upregulated after 24h.
We sought to determine whether inactivation of Notch signaling leads to differentiation of other neurons such as cone photoreceptors, another cell type generated early in development. Therefore, we analyzed additional sets of DAPT-treated retinal explants at E4.5 for changes in the cone-specific marker, Visinin (Yamagata et al., 1990; Bruhn and Cepko, 1996; Bradford et al., 2005). We found that inhibition of Notch signaling caused a dramatic increase in Visinin immunolabeling (Fig 3 A, B). We used QPCR to quantify the changes in expression of both Visinin and retinoid X receptor-□ (RXR-□), an additional early marker for cones in chick (Hoover et al., 1998). After 12h of DAPT treatment, RXR-□ showed a small, but significant increase in expression, and by 24h both Visinin and RXR-□ are uprgegulated by ∼20- and ∼15-fold respectively (Fig 3 C, D, compared in F).
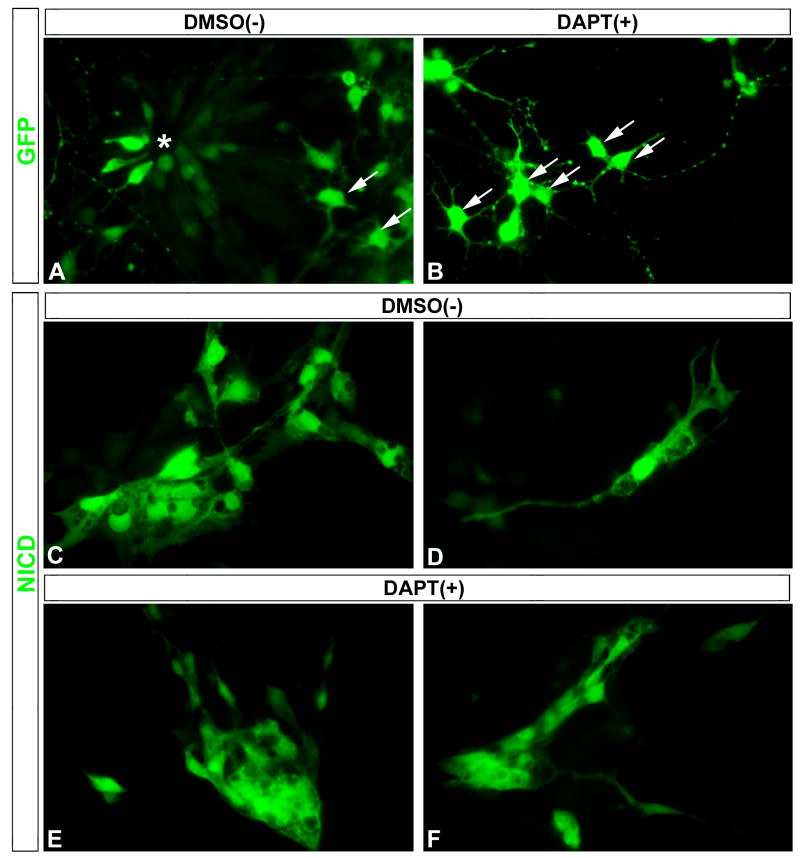
Constitutively active NICD prevents DAPT-induced neuronal differentiation
Although APP and Notch are the major substrates of the presenilin/γ -secretase complex, other type I transmembrane proteins have also been shown to be substrates for RIPping (Medina and Dotti, 2003). To determine if the effects of DAPT are specific to presenilin/γ -secretase-mediated cleavage of Notch in embryonic retinal progenitor cells, we tested whether constitutively expressed NICD could prevent the ability of DAPT to induce their differentiation. E5.5 chick retinas were dissociated and transfected with a constitutively active NICD-IRES-GFP plasmid (Daudet and Lewis, 2005) or GFP control plasmid and cultured overnight. DAPT and DMSO were added to each condition and cultured an additional 48h. In GFP-transfected cultures with the DMSO vehicle added, we observed a mix of progenitor cells and differentiating neurons typical of dissociated embryonic chick retinas (Fig 4 A). DAPT treatment of GFP-transfected cultures resulted in loss of cells with progenitor morphology and an increase in cells with neuronal appearance (Fig 4 B). NICD transfection resulted in clusters of cells with undifferentiated morphologies typical of progenitors (Fig 4 C), or often isolated cells with differentiated Muller glia-like morphology (Fig 4 D) in cultures treated with the DMSO control. Moreover, DAPT-treatment was not able to induce neuronal differentiation in NICD-transfected cells (Fig 4 E, F) as it did with GFP-transfected cells. Thus, NICD prevented DAPT-induced neuronal differentiation, supporting the notion that Notch is the major substrate of the presenilin/γ -secretase complex responsible for the effects we observe on retinal differentiation.
Figure 4. Constitutively active NICD prevents DAPT-induced neuronal differentiation.
(A, B) DAPT induced differentiation of neuronal progenitors in the GFP-transfected cells compared to control DMSO cultures: note the appearance of a large cluster of GFP+ progenitor cells found in a rosette (A, asterisk), surrounded by a few GFP+ differentiated neurons (A, arrows), while many GFP+ differentiated neurons were present in DAPT-treated cultures (B, arrows). (C–F) Transfection of NICD resulted in the appearance of large clusters of undifferentiated progenitor-like cells (C), or often isolated cells with morphologies consistent with differentiating Muller glia (D) in DMSO-treated control cultures. In contrast to the many well-differentiated neurons in DAPT-treated GFP control cultures (B), DAPT-treated NICD cultures still contained large clusters of NICD+ undifferentiated progenitor-like cells (E, F).
Notch activity regulates early and late retinal progenitors
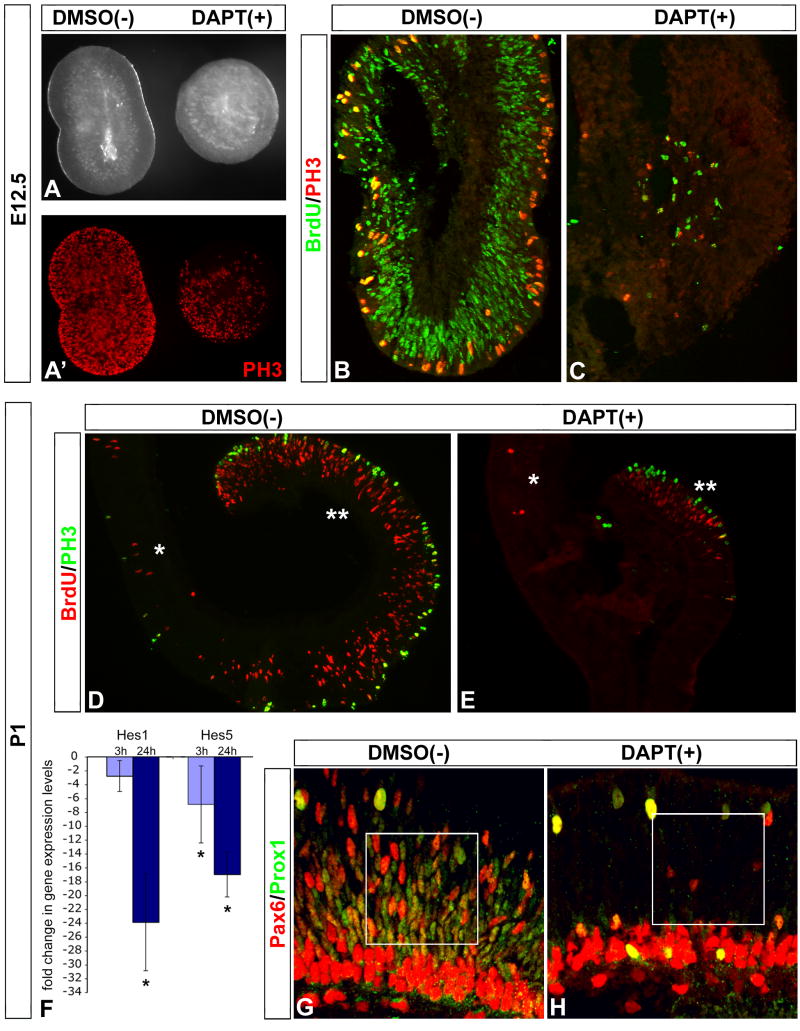
To better determine whether DAPT-mediated inactivation of Notch activity generated age-appropriate cell types, we analyzed its effects at early and late stages of mouse retinogenesis, since this process occurs over a much shorter timescale in chick. Two conditional genetic approaches based on the floxed Notch1 mouse (Radtke et al., 1999) were recently used to reduce Notch1 expression during mouse retinal development (Jadhav et al., 2006; Yaron et al., 2006). Both studies report that genetic removal of Notch1 in the early retina results in smaller eyes due to premature progenitor cell differentiation into primarily cone photoreceptors (Jadhav et al., 2006; Yaron et al., 2006). Jadhav et al (2006) also reported that removal of Notch1 later in development caused an increase in rod photoreceptors and a decrease in Muller glia. Thus we sought to determine to what extent pharmacological inactivation of Notch activity can recapitulate the phenotype observed by the genetic deletions.
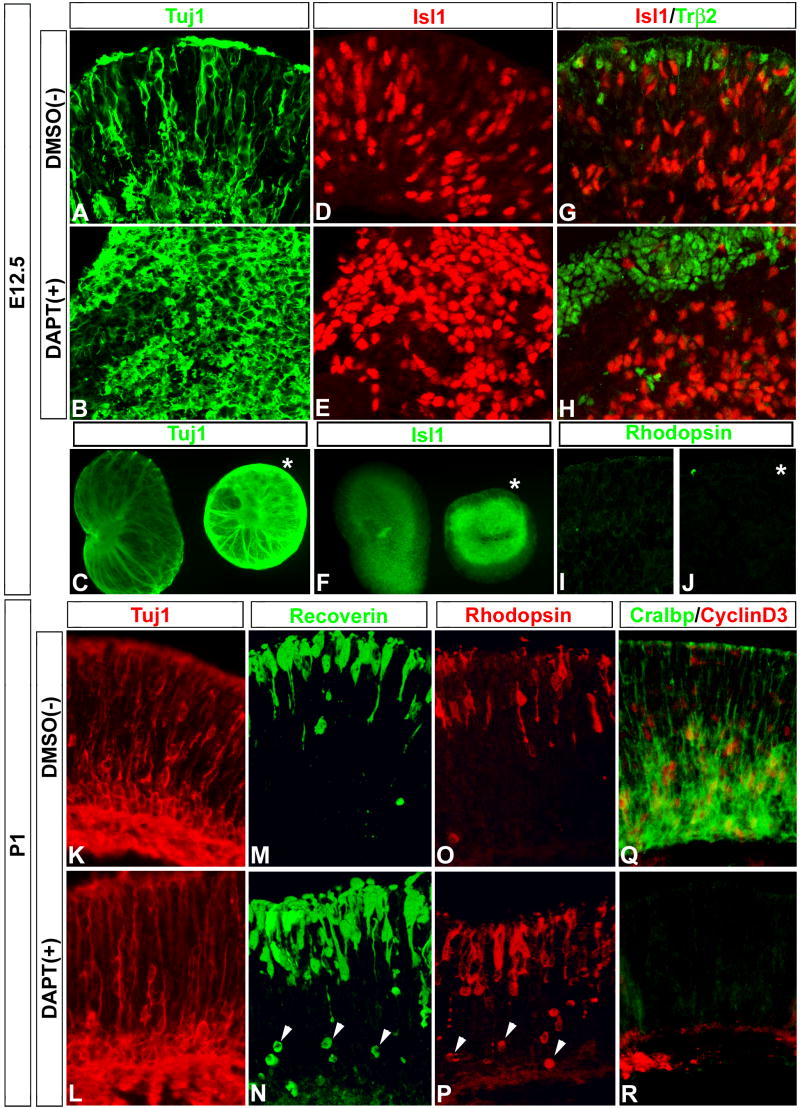
Pairs of retinas from E12.5 mice were treated with DAPT for 48h (the sister retina served as the DMSO vehicle control). Gross morphological observations of the retina pairs indicated that DAPT treatment caused a reduction in size as compared to its sister control retina (Fig 5 A). Proliferation of progenitor cells was inhibited by DAPT; both PH3 immunolabeling and BrdU incorporation were reduced in DAPT-treated retinas (Fig 5 A'–C). A similar analysis of explants from postnatal day 1 (P1) retinas also demonstrated that explants treated with DAPT were smaller and had substantially reduced BrdU- and PH3-labeled progenitor cells (Fig 5 D, E). QPCR analysis indicated that DAPT treatment resulted in a significant reduction of Hes5 gene expression levels by as early as 3h, and that by 24h both Hes5 and Hes1 transcripts declined (Fig 5 F). Additionally, fewer progenitor cells in the neuroblast layer were labeled with Pax6 and Prox1 in DAPT-treated retinas (Fig 5 G, H).
Figure 5. Notch signaling regulates proliferation of early and late retinal progenitor cells.
(A–C) E12.5 pairs of mouse retinas were cultured with DAPT or DMSO for 48h. The DAPT-treated retina was noticeably smaller than its sister control (A), and had less PH3+ progenitor cells (A'). Explants were pulsed with BrdU for 1h prior to fixation, sectioned and analyzed for BrdU incorporation and PH3 immunolabeling. Note that DAPT treatment drastically reduced the number of BrdU+ and PH3+ progenitors. (D, E) Similar analysis of pairs of P1 mouse retinal explants reveals DAPT treatment resulted in reduced retinal size, and number of BrdU+ and PH3+ progenitor cells; single asterisk marks central retina, double asterisk marks peripheral retina. (F) QPCR analysis of DAPT-treated explants resulted in a significant decrease in Hes5 gene expression levels by 3h, and both Hes5 and Hes1 levels by 24h. (G, H) DAPT treatment at P1 also produced a clear decrease in Pax6/Prox1 +/+ retinal progenitor cells (H, boxed area) compared to control (G, boxed area).
Inhibition of Notch signaling with DAPT in mouse retinas caused an increase in neuronal differentiation. There was an increase in both ganglion cell-and cone-specific markers (Tuj1 and Isl1 Fig 6 A–F, Trß2 Fig 6 G, H, respectively) in E12.5 DAPT-treated retinas, compared with control retinas. The effects of DAPT treatment on neuronal differentiation in the E12.5 retina were confined to those cell types generated early in development: we found no labeling in either control or DAPT-treated explants for later developing rod photoreceptor-specific markers (Fig 6 I, J respectively). We compared the response of E12.5 retina with that of P1 retinas. Analysis of P1 explants treated with DAPT indicated that there was no change in markers of early neuronal types such as Tuj1 (Fig 6 K, L). By contrast, later neuronal types, such as rod photoreceptors, showed a clear increase in Recoverin and Rhodopsin immunolabeling (Fig 6 M–P).
Figure 6. Notch signaling inactivation promotes stage appropriate neuronal differentiation.
(A–F) DAPT treatment of E12.5 mouse retina increased ganglion cell differentiation, visualized by Tuj1 and Islet1 (Isl1) immunolabeling of sections (A, B and D, E) and wholemounts (C) and (F) respectively: asterisk in (C) and (F) mark DAPT-treated explant. (G–J) DAPT treatment at E12.5 also increased cone photoreceptor differentiation visualized by Trß2 immunolabeling (G, H), but not later-born rod photoreceptors visualized by lack of Rhodopsin immunoreactivity in both control and DAPT-treated explants (I, J respectively). (K–R) DAPT treatment at P1 had no effect on earlier-born Tuj1+ neurons (K, L), but produced a clear increase in both Recoverin and Rhodopsin immunolabeling of rod photoreceptors in the apical region of the outer nuclear layer (M–P); also note the numerous newly generated rods in the more basal region of the outer nuclear layer (arrowheads, N, P). DAPT treatment also produced a clear decrease in the differentiation of Muller glia cells, which have just begun to differentiate as visualized by CRALBP and Cyclin D3 immunolabeling (Q, R).
Others have shown that effectors of the Notch signaling pathway, such as Hes1, Hes5, and Hey2, are important for generating Muller glia cells (Tomita et al., 1996; Furukawa et al., 2000; Hojo et al., 2000; Satow et al., 2001; Takatsuka et al., 2004), and that conditional deletion of Notch1 in late retinal clones resulted in a reduction of Muller glia (Jadhav, et al 2006). We found that two Muller glial markers, CRALBP and CyclinD3, were reduced in the DAPT-treated retinas (Fig 6 Q, R). Thus, DAPT treatment at both early and late stages of mouse retinal development reduced retinal size, the number of progenitor cells, and Hes5 and Hes1 expression levels, in a manner similar to that in the chick. DAPT treatment also initiated differentiation of neuronal cell types specific to the stage at which they are normally generated, and inhibited development of Muller glia.
Transient inactivation of Notch signaling initiates permanent neural differentiation
It has been reported that a transient activation (24h) of Notch signaling causes a permanent switch in cultured neural crest stem cells to undergo gliogenesis rather than neurogenesis (Morrison et al., 2000). To determine whether a transient inactivation of Notch signaling can commit progenitor cells to neural differentiation, we exposed developing retinas to progressively longer periods of DAPT treatment. E4.5 chick retinas were bisected and one half of the explant treated with DAPT for 1h, 3h, 6h, 12h, 24h, and 48h, while the other half of the explant served as a time-matched DMSO control. After the period of DAPT exposure, the explants were washed three times and cultured in DAPT-free media for a total of 48h. They were then fixed and immunolabeled with antibodies to PH3 and Visinin, and analyzed by LSCM as described above.
While DAPT treatment for 1h or 3h did not alter retinal development, periods of DAPT treatment for 6h or longer produced a clear effect on retinal development (Sup Fig 2). Inactivating Notch signaling for 6h caused a noticeable reduction in size, and this became more apparent with longer exposures to DAPT (Fig 7 A). Permanent changes in progenitor cell proliferation occurred from periods of 6h or more of DAPT treatment, and large regions devoid of PH3+ progenitors cells were observed (Fig 7 B–D; Sup Fig 2). There was a concurrent increase in Visinin immunolabeling in cultures treated with DAPT for longer than 6h (Fig 7 C, D, and data not shown).
Figure 7. Transient inactivation of Notch signaling synchronizes neural differentiation.
(A) Bisected E4.5 chick retinal explants were exposed to DAPT for 1h, 3h, 6h, 12h, 24h, washed extensively and cultured in DAPT-free media for a total of 48h. Explants exposed to periods DAPT treatment for 6h were noticeably smaller in size than their control, which became more apparent with increased exposure to DAPT (asterisks). (B–D) Explants were wholemount immunolabeled with antibodies to PH3 and Visinin, and analyzed by LSCM as before. Exposure to DAPT for periods of 6h or longer produced a clear reduction in PH3+ progenitor cells compared to controls, with an concomitant increase in Visinin immunoreactivty over time. No change in the pattern of PH3 or Visinin immunolabeling was observed in periods of DAPT exposure less than 6h (Sup Fig 2, 3).
We also found a consistent spatial sensitivity to the transient inactivation of Notch signaling. Progenitor cells located in the central region of the retina were more sensitive to a transient decrease in Notch activity, while longer exposures to DAPT were required to commit more peripheral progenitor cells to differentiate. After 6h of DAPT treatment, there was a boundary between the differentiating central retina and the seemingly normal peripheral region, which became more apparent after 12h of DAPT treatment (Sup Fig 2, 3). However, with 24h of DAPT treatment, even peripheral regions differentiated (data not shown).
Synchronized Notch signaling inactivation initiates a proneural bHLH cascade leading to differentiation
Although Hes5 gene expression was decreased by 3h of Notch signaling inactivation, the above results demonstrate that a minimum of 6h was required for progenitor cells to permanently commit to differentiation. Proneural bHLH genes are known to be direct targets of the Notch effectors, and are important for proper neuronal differentiation (reviewed by Fisher and Caudy, 1998; Bertrand et al., 2002; Quan and Hassan, 2005). We hypothesized that a critical threshold of de-repression of proneural bHLH genes must be achieved to permanently commit progenitor cells to differentiation.
We compared the temporal changes in expression of Cash1, Ngn2, NeuroM, NeuroD, and Cath5 in DAPT-treated E4.5 chick retinal explants to that of controls by QPCR. Comparing the relative changes of these genes revealed a dynamic set of expression profiles that fell into three sequentially and transiently upregulated groups. (1) Cash1 and Ngn2 were upregulated by 3h, and reached their peak expression levels at 6h and 12h respectively: both were downregulated to below untreated levels by 48h (Fig 8 A, B respectively). (2) NeuroM expression was increased at 6h, reached its peak expression by 12h, and decreased to untreated levels by 48h (Fig 8 C). (3) NeuroD and Cath5 did not show increases until 12h; Cath5 levels declined to those of untreated retinas by 48h, while NeuroD remained elevated (Fig 8 D, E respectively). These results support the possibility that a critical threshold in either Cash1 or Ngn2 might be reached within 6h of inhibition of the Notch pathway, thereby committing the progenitor cells to terminal differentiation (Fig 8 F). Moreover, these results are also consistent with the possibility that the various bHLH genes in the retina are activated in a cascade, with the group 1 genes, like Cash1 and Ngn2 activating “downstream” bHLH genes, like NeuroM and Cath5.
Figure 8. Synchronized Notch signaling inactivation reveals an inherent proneural bHLH cascade during progenitor cell differentiation.
QPCR was used to analyze changes in E4.5 chick retinal explant pairs treated with DAPT compared to control over time as before. Cash1 (A), Ngn2 (B), NeuroM (C), NeuroD (D), and Cath5 (E) all exhibit statistically significant dynamic temporal changes over time due to Notch signaling inactivation. (F) Comparing the relative patterns of changes reveal that three distinct transient and sequential responses: (1) Cash1 and Ngn2 were transiently upregulated at 3h, with Cash1 peaking at 6h and Ngn2 peaking at 12h, and both downregulated to below normal levels by 48h; (2) NeuroM transiently increased later at 6h, peaked at 12h, and decreased to normal levels by 48h; (3) NeuroD and Cath5 increased even later sometime after 6h, with both peaking at 12h: Cath5 decreased back to normal levels while NeuroD remained elevated by 48h (Fig 7 D, E respectively).
Differential Notch activity within individual retinal progenitors
Transient manipulations of Notch activity, either inactivating (this report) or activating (Morrison et al., 2000), suggest that brief alterations in the levels of Notch activity within individual progenitor cells can determine whether or not a progenitor cell differentiates. Our results show that only a relatively short period of Notch inactivation (6 hr) is sufficient to commit a progenitor to differentiate, and suggest a model in which fluctuations in the level of Notch signaling in progenitors underlies the normal mechanism for differentiation. Tokunaga and colleagues demonstrated that different levels of activated Notch are observed in progenitor cells in the nervous system during development (Tokunaga et al., 2004), although retinal expression was not reported. To determine if retinal progenitor cells exhibit different levels of activated Notch signaling, we used an antibody specific to the γ-secretase-mediated cleavage product NICD (ActN1, as in Tokunaga et al., 2004). At E14.5, ActN1 is confined to the neuroblast layer of the developing retina, and restricted from the ganglion cell layer and peripheral regions where the ciliary body and iris would be located (Fig 10 A). Within the neuroblast layer, ActN1 is not expressed in the differentiating neurons (TuJ1+), but only in retinal progenitor cells: higher levels of ActN1 are observed in S-phase progenitor cells and lower levels of ActN1 are observed in M-phase progenitor cells (Fig 10 F–J), similar to progenitor cells elsewhere in the nervous system (Tokunaga et al., 2004). These data show that Notch signaling activity changes during the cell cycle, reaching a low point during M-phase.
Figure 10. Notch signaling activity fluctuates in mitotic retinal progenitor cells.
E14.5 mouse retinal sections were immunolabeled with an antibody specific to the activated form of NICD (ActNotch1, purple cells) to determine the pattern of Notch activity in situ (Tokunga et al., 2004). (A, B) Notch activity is observed in the neuroblast layer of the retina (brackets), but not in the ganglion cell layer (gc) or in peripheral regions where the ciliary body and iris would be located (A, arrows). Also, note that subpopulations of cells are more heavily labeled than others (arrowheads; A, transverse section; B, oblique transverse section). (C–E) ActNotch1 immunolabeling (arrows) is not observed in Tuj1+ differentiating neurons (red cells, arrowheads). (F–J) To observe Notch activity within retinal progenitor cells, sections from an E14.5 mouse embryo that received a 1h pulse of BrdU in utero prior to sacrifice were sequentially processed for actNotch1 staining, immunolabeling for PH3 and BrdU incorporation, and counterstaining with Dapi. Panels are arranged to show (F) actNotch1 (purple), (G) actNotch1 and PH3 (red), (H) actNotch1 and BrdU (green), (I) actNotch1, PH3, and BrdU, (J) PH3, BrdU, and Dapi. (F'–I') Insets from F–I are shown at higher magnification respectively. (F"–H", J") Same insets are shown overlayed onto Dapi to faciltate comparison: actNotch1+/BrdU+ progenitor cells are marked with asterisks; PH3+ progenitor cells at the mitotic surface are marked with arrowheads. Higher levels of activated Notch are observed in S-phase progenitor cells compared to M-phase progenitor cells.
Synchronized Notch signaling inactivation reveals new components in the initial program of progenitor cell differentiation
To determine the scope of molecular changes during the initial phase of Notch signaling inactivation, we compared global gene expression of E14.5 mouse retinal explants treated with DAPT for 8h, to that of controls, using microarray analysis (Affymetrix Mouse Genome 430 2.0 Array, Sup Fig 4). We used QPCR to validate changes in expression levels of selected genes from the array (Fig 11). The microarray/QPCR analysis confirmed that Hes1 and Hes5 are downregulated with DAPT treatment (Fig 11; Sup Fig 4). By contrast, the proneural bHLHs Mash1, Ngn2, NeuroD1, and Math5 were upregulated in DAPT-treated retinas (Fig 11; Sup Fig 4). Additionally, microarray/QPCR analysis identifies changes in expression levels of other members of the Hes and proneural bHLH families: Idb3, Idb4, and Dtx4 are downregulated while Hes6 is upregulated; Bhlhb5 is upregulated while Bhlhb2 is downregulated (Fig 11; Sup Fig 4). The upregulation of Bhlhb5 is intriguing, as it has recently been shown to regulate amacrine and cone bipolar formation (Feng et al., 2006). Thus at E14.5, an increase in Bhlhb5 expression would likely correlate with increased amacrine differentiation, further demonstrating that Notch signaling also regulates the genesis of this cell type in the early retina. Expression of the Notch ligands Dll1 and Dll4 are upregulated (Fig 11). Thus, DAPT-treatment causes a coordinated response amongst Notch signaling pathway components, including Notch effector genes, proneural bHLH transcription factors, and Notch ligands (Fig 11, Sup Fig 4). Furthermore, QPCR confirms the majority of changes observed by microarray analysis, indicating a high degree of correlation between the two methods.
Figure 11. Identification of new components involved in the initial program of progenitor cell differentiation.
E14.5 mouse retinal explants were treated with DAPT or DMSO for 8h, and global changes in gene expression due to DAPT treatment were compared by microarray analysis (Affymetrix Mouse Genome 430 2.0; Supp Fig 4). QPCR was used to confirm changes in expression levels of selected genes identified from the microarray analysis as before: note that similar genetic changes are observed at this early timepoint in mouse and chick (Hes and proneural bHLH genes). Additionally, microarray/QPCR analysis identifies changes in other signaling systems such as FGF, Wnt, and IGF pathways, changes in other gene family members such as Delta 1/4 and Hes6, and finally previously uncharacterized genes during retinal development such as Insulinoma-associated 1 (Insm1).
Changes in genes associated with other signaling pathways were also observed: Fgf3, 13, and 15, the Wnt inhibitors Sfrp2 and Dkk3, and insulin growth factor binding proteins Igfbp 1,4, all showed decreased expression by 8h of DAPT treatment (Fig 11). Chx10 and Rax, homeodomain transcription factors associated with retinal progenitor cells, already indicate reduced gene expression levels by 8h of DAPT-treatment (Fig 11). Additionally, changes were observed in transcription factors and/or DNA-binding proteins previously not characterized as regulated by Notch input during retinal development. Notably, Nr2e1 (Tlx), an orphan nuclear receptor known to be essential for retinal progenitor cell proliferation (Miyawaki et al., 2004; Zhang et al., 2006), is substantially downregulated by 8h of Notch signaling inactivation. Conversely, Sox4 and Sox11 have begun to be upregulated by this time (Fig 11), consistent with possibility that some Sox family members function to promotte progenitor cell differentiation, such as that observed in the spinal cord with Sox1–3 and Sox21 activities (Bylund et al., 2003; Sandberg et al., 2005). Repressor protein 58 (RP58) and insulinoma-associated 1 (Insm1) are zinc-finger proteins that are upregulated due to Notch signaling inhibition (Fig 11). RP58 is a DNA-binding protein that mediates sequence-specific transcriptional repression from E-box motifs, is associated with heterochromatin, and recruits a corepressor complex with Dnmt3a methylase and HDAC1 histone deacetylase (Aoki et al., 1998; Meng et al., 2000; Fuks et al., 2001). Insm1 is a transcription factor necessary for endocrine cell differentiation in the pancreas (Gierl et al., 2006), and is regulated by NeuroD1 and Ngn3 (Breslin et al., 2003; Mellitzer et al., 2006); its function during retinal development is not known. Finally, many components of the cell cycle machinery were observed to change after 8h of Notch signaling inactivation (data not shown), two of which were Btg2 and CyclinD1. Btg2 expression increased after DAPT treatment, and its activity is linked to increased lengthening of the cell cycle and progression toward neuronal differentiation (Iacopetti et al., 1999; Calegari et al., 2005). A slightly increased level of CyclinD1 was also observed, although this would be the opposite of what would be predicted upon synchronized differentiation. However, as many other cell cycle components also showed increased or decreased expression levels as well, it remains to be determined how the Notch signaling pathway and the cell cycle machinery intersect.
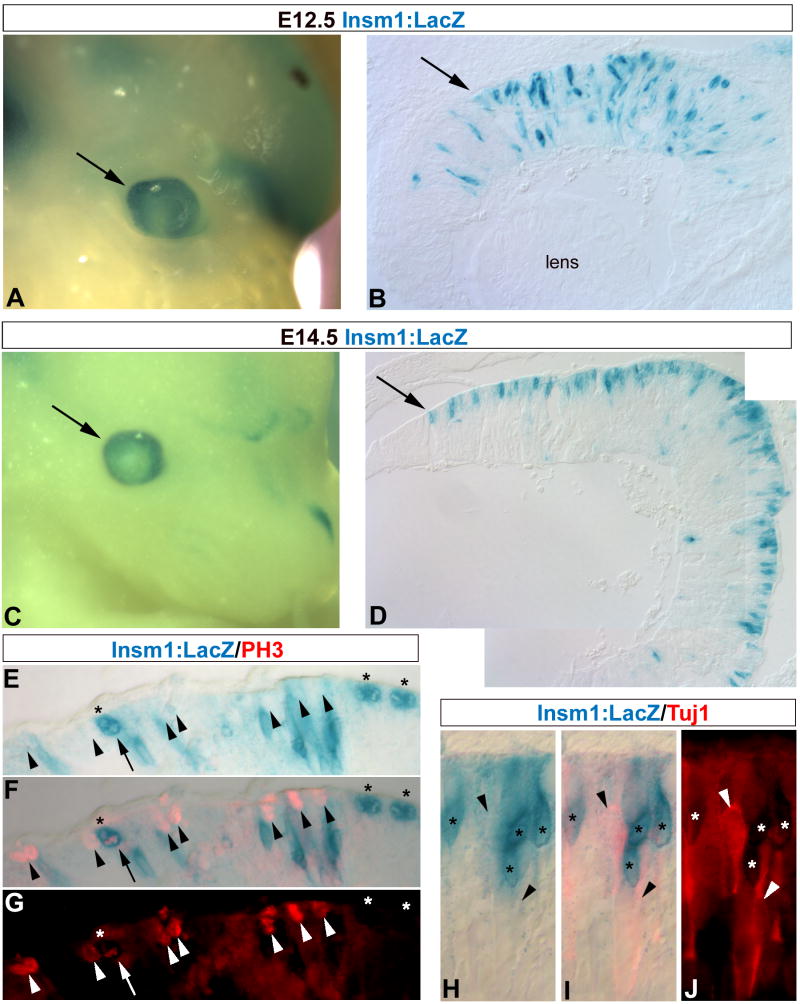
To validate this approach for the identification of novel components of the neuronal differentiation pathway, we analyzed the expression of Insm1, a zinc-finger transcription factor regulated by proneural bHLH transcription factors. Insm1 has been shown to mediate differentiation of newly born endocrine cells in the pancreas (Breslin et al., 2003; Gierl et al., 2006; Mellitzer et al., 2006) and a transgenic Insm1:LacZ reporter mouse has been generated (Breslin et al., 2003). We used this mouse line to determine what cell type(s) express Insm1 during retinal development. Insm1:LacZ reporter is expressed in a discrete population of cells in the central retina at E12.5 (Fig 12 A, B). By E14.5, Insm1:LacZ is primarily restricted to cells located at the ventricular surface, although an occasional cell is observed in the ganglion cell layer (Fig 12 C, D). PH3 immunolabeling reveals that the majority of Insm1:LacZ+ cells at the ventricular surface are not dividing progenitor cells (Fig 12 E–G) or Tuj1+ differentiating ganglion cells migrating to the ganglion cell layer (Fig 12 H–J), although one Insm1:LacZ+/PH3+ cell was observed. Therefore, Insm1 is likely expressed very early during differentiation, most likely in newly born photoreceptors at this age, which have previously been shown in this layer (Hinds and Hinds, 1979; Furukawa et al., 1997; Roberts et al., 2005).
Figure 12. Insulinoma-associated 1 (Insm1) is expressed early during retinal development.
E12.5 and E14.5 Insm1-LacZ transgenic mice were used to characterize Insm1 expression during early retinal development (Lan et alreference). Insm1:LacZ activity is detected in the developing eye early at E12.5 (A, arrow). Sections reveal Insm1:LacZ expression in a discrete subpopulation of cells distributed through the central retina (B, arrow). By E14.5 Insm1:LacZ expression in the eye (C, arrow) is primarily confined to the ventricular surface (D, arrow). (E–G) PH3 immunolabeling reveals that at this stage (E14.5) the majority of PH3+ mitotic progenitor cells (red, arrowheads) are not Insm1:LacZ+ (blue, asterisks), although one Insm1:LacZ+/PH3+ cell was observed (arrow). (H–J) However, Tuj1 immunolabeling at this stage reveals that Insm1:lacZ+ cells (asterisks) in this outer layer are also not Tuj1+ (red, arrowheads).
Discussion
In this report, we show that pharmacological inhibition of Notch signaling can phenocopy the experimental results obtained with other methods, but allows for better temporal control over the differentiation process. Treatment of developing retina with DAPT causes the following: 1) a rapid decline in downstream components of the Notch signaling pathway that initiates a molecular cascade leading to synchronized differentiation of progenitors; 2) a stage dependent differentiation of the various retinal cell types; 3) a permanent commitment to differentiation after transient exposure; and 4) an inherent cascade of proneural bHLH gene expression underlying the entire process. Thus, DAPT provides a powerful tool for the synchronization of the cell differentiation processes regulated by Notch activity.
DAPT recapitulates genetic manipulation of Notch signaling pathway components
Deletion of Notch1 causes early embryonic lethality prior to retinal development (Conlon et al., 1995; de la Pompa et al., 1997), but recently two studies have reported the effects of a Notch1 conditional knockout (CKO). These mice have smaller eyes, reduced progenitor cell proliferation, and increased differentiation of cone photoreceptors early (Jadhav et al., 2006; Yaron et al., 2006) and rod photoreceptors later (Jadhav et al., 2006). We report that DAPT treatment has similar effects: the DAPT-treated retinas are smaller, have decreased proliferation, and increased neuronal differentiation. DAPT also causes premature differentiation of cone photoreceptors in embryonic retina, and differentiation of rod photoreceptors in postnatal retina. In addition, both Notch1 CKO (Jadhav et al., 2006) and DAPT treatment result in an inhibition of Muller glia differentiation. Thus, the effects of DAPT treatment are consistent with, and confirm, the results of the Hes1, Hes5, and the Notch1 CKO genetic studies.
However, there is one main difference between the Notch1 CKO studies and our results with DAPT: DAPT treatment causes an increase in ganglion cell differentiation that was not observed in either Notch1 CKO study. This discrepancy may be due in part to the timing and variability of expression of the Chx10-Cre driver from one study (Rowan and Cepko, 2004; Jadhav et al., 2006), or the α-Pax6-Cre driver from the other study (Yaron et al., 2006) used to conditionally delete Notch1 in the retina. The difference may also be due to redundancy between Notch family members: both Notch1 and Notch3 are expressed in the early neural retina (Lindsell et al., 1996). DAPT treatment caused a large reduction in Hes5 and Hes1 expression, as did the α-Pax6-Cre Notch1 CKO (Yaron et al., 2006), but the Chx10-Cre Notch1 CKO did not (Jadhav et al., 2006). An analogous study in the cortex demonstrating functional redundancy between Notch1 and Notch3 was accompanied by loss of Hes5 and Hes1 in the retina (FoxG1-Cre, Mason et al., 2005). Our results in the developing chick and mouse retina are also somewhat different from those in zebrafish (Bernardos et al 2005). A different γ -secretase inhibitor (Compound E) caused a disruption in lamination, a change in cone spectral subtype, and an inhibition of Muller glia development, but neither mindbomb mutation nor Compound E caused a premature depletion of the progenitor pool (Bernardos et al., 2005). The difference between fish and other vertebrates may be due to a difference in the rate of development. Rapidly developing systems may not rely on Notch signaling for maintaining their progenitor pool, whereas systems requiring prolonged times for development are more sensitive to this aspect of Notch signaling activity. Nevertheless, DAPT treatment phenocopies various aspects of other Notch pathway mutations in zebrafish and Drosophila (Geling et al., 2002; Micchelli et al., 2003).
It is possible that some of the effects we observe with DAPT are due to inhibition of other presenilin/γ-secretase substrates. However, we consider this unlikely for several reasons. First, overexpression of the NICD in retinal progenitors prevented their DAPT-mediated differentiation (Fig 4). Second, many of the known components of the Notch signaling pathway changed in predictable ways due to DAPT treatment (Sup Fig 4, Fig 11). Third, we did not observe a change in most of the target genes from presenilin/γ-secretase substrates other than the Notch pathway (eg. APP and the amyloid precursor-like protein APLP1; Hebert et al., 2005). Although, we observed a small decrease in APLP2 and an increase in GSK3ß expression (one of the putative targets of APLP2; Xu et al., 2006), these changes were in the opposite direction of what would be predicted by inhibition of APLP2 processing.
Transient inhibition of Notch activity permanently commits progenitor cells to differentiate
While previous studies of neural crest stem cells have shown that exposure to an activating Notch signal for 24h irreversibly committed these cells to glia (Morrison et al., 2000), the period of Notch inactivation needed for irreversible commitment to differentiation was not known. Our experiments show that less than 6h of DAPT exposure allows the progenitors to recover and remain in the cell cycle, but periods of DAPT treatment longer than this lead to differentiation. It is not clear why 6h is the critical time for Notch inactivation to commit progenitors to differentiate, since Notch activity is down-regulated after only 3h (summarized in Fig 9).
Figure 9. Summary of changes in cellular and molecular kinetics due to loss of Notch signaling.
Cycling progenitor cells that receive a pulse of DAPT for 3h or less are able to recover and remain progenitors (A, white to gray), even though Notch effector genes have been downregulated (B). However exposure to periods of DAPT treatment for 6h or longer causes a permanent commitment to differentiate (large arrows). Committed progenitor cells increase expression of general differentiation genes by 12h, with a concomitant decrease in expression of progenitor genes and an increase in expression of cell-type specific genes by 24h. By 48h after Notch signaling inactivation, decreased proliferation and widespread neuronal differentiation are readily apparent. If Notch signaling is inactivated in early in development, then early cell types such as cones and ganglion cells are generated (as depicted). If Notch is inactivated during late retinal development, then increases in later cell types are observed, such as rods, along with conconmittent decreased Muller glia differentiation. The inherent cascade of proneural bHLH transcription factors initiated by synchronized Notch signaling inactivation underscores the temporal requirement for progenitor cells to permanently commit to neuronal differentiation.
One possibility may relate to the observations that Notch is normally active only during the S-phase of the cell cycle, and not during M-phase (Fig 10; Tokunaga et al., 2004). If substantially shorter periods of Notch inactivation were required to commit cells to differentiate, the cells would not have sufficient time for the mitotic phase of the cycle. The length of M-phase may thus serve as a limit to the length of time during which Notch can be inactive, yet still preserve the cell in an undifferentiated state. While this does not explain why Notch activity oscillates with the cell cycle, it may explain why preventing cells from exiting M-phase promotes their differentiation (Murciano et al., 2002).
An alternative explanation for the minimum 6h requirement is that another factor reaches a critical threshold at this time. Since the proneural bHLH genes are immediate targets of Hes1/5, they seemed likely candidates for this role. Indeed, Cash1, Ngn2, and NeuroM show significant increases in expression after 6h of DAPT treatment (summarized in Fig 9). Low level expression of proneural bHLH genes is necessary for expression of Notch pathway components in neural progenitor cells; eg. Delta1 (Fode et al., 1998; Ohsawa et al., 2006) and Hes1/5 (Castro et al., 2005; Lamar and Kintner, 2005). However, overexpression of Mash1 or Ngn2 promotes cell cycle withdrawal, migration away from the ventricular zone, and neuronal differentiation (Simmons et al., 2001; Novitch et al., 2001; Bylund et al., 2003; Lee and Pfaff, 2003; Sandberg et al., 2005; Helms et al., 2005; Fior and Henrique, 2005). Moreover, Tokunaga et al (2004) found that forebrain progenitor cells expressing high levels of either Ngn2 or Mash1 had the lowest levels of ActN1 (Tokunaga et al., 2004). Therefore, while neural progenitor cells normally express proneural bHLH genes, increased expression beyond a threshold level could commit them to differentiate.
As noted above, the timing of changes in expression of the bHLH transcription factors in both chick and mouse following DAPT treatment is consistent with a cascade in their function. Cash1 and Ngn2 were upregulated by 3h, while NeuroM expression was upregulated after 6 hours and NeuroD and Cath5 did not show increases until 12h. It has been proposed that proneural bHLH transcription factors operate as a cascade, whereby upstream bHLHs in progenitor cells induce downstream bHLHs to promote neural differentiation. Intriguingly, there is evidence that Mash1/Ngn2 induce such cascades in the olfactory epithelium, spinal cord, and somewhat in the retina (Cau et al., 1997; Novitch et al., 2001; Lee and Pfaff, 2003; Bylund et al., 2003; Skowronska-Krawczyk et al., 2004; Sandberg et al., 2005; Fior and Henrique, 2005; Matter-Sadzinski et al., 2005). It is striking that such a temporally distinct and dynamic cascade of bHLH gene expression is observed after synchronized Notch inactivation, which allowed us to more comprehensively visualize this cascade in the retina for the first time.
Synchronizing progenitor cell differentiation identifies new components of the differentiation program
Microarray/QPCR analysis of DAPT treated retinas allowed us to identify new components of the differentiation program of neural progenitor cells. Characterization of Insm1, one of the genes upregulated after 8h of DAPT treatment, validates our approach to discover new pathways operating during initial stages of progenitor cell differentiation. Insm1 is particularly interesting in regard to its function during endocrine cell differentiation in the pancreas and gut. Deletion of Insm1 completely stalls the differentiation of endocrine precursor cells (Gierl et al., 2006). Proneural bHLH genes Ngn3 and NeuroD1 regulate Insm1 expression in endocrine precursor cells (Breslin et al., 2003; Mellitzer et al., 2006), and Insm1 seems to feedback to repress NeuroD1 activity (Liu et al., 2006). Analysis of retinas from Insm1:LacZ reporter mice reveals that Insm1 is likely expressed in newly differentiating photoreceptors at this age (Fig 12). It will be interesting to determine whether Insm1 has a similar role in promoting downstream events in differentiating photoreceptors, and how proneural bHLH genes regulate Insm1 expression. Among the bHLH transcription factors that were upregulated after DAPT treatment was Bhlhb5. Gan and colleagues recently reported that Bhlhb5 is required for the differentiation of amacrine cells and subtypes of cone bipoplar cells (Feng et al., 2006). Increased expression of Bhlhb5 in our experiments at E14.5–E15.5 likely reflects increased amacrine cell differentiation (Fig 11). Thus, this approach also demonstrates that synchronized Notch signaling inactivation reveals molecular changes associated with the differentiation of age appropriate cell types. Our approach also revealed a Notch signaling input in Fgf, Wnt, and insulin signaling pathways (Fig 11). The mechanism through which Notch signaling regulates these diverse pathways remains to be elucidated. Further analysis of additional time-points should provide more information about the temporal dynamics of the molecular program of progenitor cell differentiation.
It has been proposed that there is a “clock” in retinal progenitor cells, likely reflecting changing competence as time goes by. Simply triggering the inactivation of Notch at progressively later stages of retinal development provides a mechanism to generate a sequence of different types of cells (Reh and Cagan, 1994; Cepko et al., 1996). Components of the Notch pathway are known to function in a “clock-like” manner (discussed by Bessho and Kageyama, 2003), although it remains unclear how a Notch regulatory “clock” intersects with the observed changes in competency of progenitor cells over time. Notch activity may act simply to reset the clock during each cell cycle, while integration of other intrinsic and extrinsic signals may regulate the competence to differentiate into a specific cell type at a given time (Li et al., 2004; Kim et al., 2005). Alternatively, Notch activity itself may progressively limit progenitor competence by a ratchet-like mechanism, such that each cell cycle results in a smaller repertoire of fate decision available to progenitors over time. Synchronization of progenitor cell differentiation should allow a systematic analysis of this process.
Supplementary Material
DAPT was added to one E4.5 chick retinal explant, while DMSO was added to the sister as control. After 6h of exposure to DAPT, propidium iodide (PI) was added and incubated for 5 min. Explant pairs were washed with cold HBSS+, and flat-mounted between glass coverslips on ice. LSCM images of DMSO control (A) and DAPT-treated explant (B) from a region between the optic nerve head and folded edge of the explant. (C) Quantification of total number of PI+ cells in a given volume reveals that there is no statistical difference between DAPT-treated explants compared to control (n=3 pairs, Student's T-test, P<0.3185).
E4.5 chick retinal explant pairs were treated with DAPT for the indicated times, washed three times, cultured for a total of 48h, and immunolabeled as wholemounts with the mitotic indicator PH3 located apically, and Islet1 (Isl1) to label ganglion cells located basally. All panels are oriented such that central-to-peripheral retina is left-to-right. Images of control explants from all ages, and DAPT-treated explants from 1h and 3h exposures are taken at the folded edge of the flattened explants as depicted: single asterisk denotes normal pattern of labeling; double-headed arrows denote the edge of the explant. No effect of DAPT exposure for 1h or 3h was observed compared to controls. However periods of 6h or longer caused an permanent decrease in PH3 immunoreactivity in a central-to-peripheral wave, concomitant with increased Isl1 immunoreactivity. Note that at 6h a clear boundary is observed between the effected region located centrally, and the apparently normal region located more peripherally (dashed line).
Transient exposure to DAPT for 6h (A) produced a consistent reduction of PH3 (green) and increased Visinin (red) immunolabeling in a central-to-peripheral pattern, such that a clear boundary (arrow) was observed between the differentiated central region (single asterisk) and apparently normal peripheral region (double asterisks), which became even more apparent with 12h exposure to DAPT (B; PH3, blue; Visinin, red). The neural retina was fully differentiated with exposures to DAPT longer than 12h (data not shown).
Affymetrix arrays were used to compare global changes gene expression between E14.5 mouse retinas treated with DAPT for 8h and controls (n=12 retinas per chip, Affymetrix Mouse Genome 430 2.0 Array). Molecular changes were ranked according to initial fluorescence intensity units (FIU, > 0.5) and examined for Notch pathway associated genes. A scatter plot depicts genes identified by these criteria, and the table contains all raw information accordingly.
Acknowledgments
We thank N. Daudet for the NICD-IRES-GFP plasmid, and O. Bermingham-McDonough, D. Raible, and P. Etter for critical comments on the manuscript. This work was supported by NRSA postdoctoral fellowship F32 EY15631-01A1 to B. R. N., and NIH grants RO1 NS28308 and EY13475 to T. A. R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121(11):3637–50. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Alexson TO, Hitoshi S, Coles BL, Bernstein A, van der Kooy D. Notch signaling is required to maintain all neural stem cell populations--irrespective of spatial or temporal niche. Dev Neurosci. 2006;28(1–2):34–48. doi: 10.1159/000090751. [DOI] [PubMed] [Google Scholar]
- Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem. 1998;273(41):26698–704. doi: 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87(7):1191–202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278(2):381–95. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3(7):517–30. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Kageyama R. Oscillations, clocks and segmentation. Curr Opin Genet Dev. 2003;13(4):379–84. doi: 10.1016/s0959-437x(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Bradford RL, Wang C, Zack DJ, Adler R. Roles of cell-intrinsic and microenvironmental factors in photoreceptor cell differentiation. Dev Biol. 2005;286(1):31–45. doi: 10.1016/j.ydbio.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278(40):38991–7. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Bruhn SL, Cepko CL. Development of the pattern of photoreceptors in the chick retina. J Neurosci. 1996;16(4):1430–9. doi: 10.1523/JNEUROSCI.16-04-01430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat Neurosci. 2003;6(11):1162–8. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25(28):6533–8. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132(15):3333–44. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124(8):1611–21. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. PNAS. 1996;93(2):589–95. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A. Why is delta endocytosis required for effective activation of notch? Dev Dyn. 2006;235(4):886–94. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyung JH, Raper DM, Selkoe DJ. Gamma-secretase exists on the plasma membrane as an intact complex that accepts substrates and effects intramembrane cleavage. J Biol Chem. 2005;280(6):4383–92. doi: 10.1074/jbc.M409272200. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–45. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132(3):541–51. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, Conlon RA. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124(6):1139–48. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Rapaport DH, Harris WA. Xotch inhibits cell differentiation in the Xenopus retina. Neuron. 1995;14(3):487–96. doi: 10.1016/0896-6273(95)90305-4. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385(6611):67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34(1):53–8. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Feng L, Xiel X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;2006;133:4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;281(2):318–33. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20(4):298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20(3):483–94. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20(10):2536–44. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91(4):531–41. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26(2):383–94. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–90. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3(7):688–94. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20(17):2465–78. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ES, Stubbs JL, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003;130(3):539–52. doi: 10.1242/dev.00275. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Reh TA. Homologies between vertebrate and invertebrate eyes. in Drosophila Eye Development. In: Moses K, editor. Results Probl Cell Differ. Vol. 37. Heidelberg: Springer; 2002. pp. 219–256. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7(7):739–45. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132(12):2709–19. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signaling in the embryonic chick retina. Curr Biol. 1997;7(9):661–70. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Differentiation of photoreceptors and horizontal cells in the embryonic mouse retina: an electron microscopic, serial section analysis. J Comp Neurol. 1979;187(3):495–511. doi: 10.1002/cne.901870303. [DOI] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127(12):2515–22. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Hoover F, Seleiro EA, Kielland A, Brickell PM, Glover JC. Retinoid X receptor gamma gene transcripts are expressed by a subset of early generated retinal cells and eventually restricted to photoreceptors. J Comp Neurol. 1998;391(2):204–13. [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner WB. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. PNAS. 1999;96(8):4639–44. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133(5):913–23. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308(5730):1927–30. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C. The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development. 2005;132(16):3619–30. doi: 10.1242/dev.01937. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38(5):731–45. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lewis J. Neurogenic genes and vertebrate neurogenesis. Curr Opin Neurobiol. 1996;6(1):3–10. doi: 10.1016/s0959-4388(96)80002-x. [DOI] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43(6):795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8(1):14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Liu WD, Wang HW, Muguira M, Breslin MB, Lan MS. INSM1 functions as a transcriptional repressor of the neuroD/beta2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem J. 2006;397(1):169–77. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S. Stem cells: You make me feel so glial. Curr Biol. 2000;10(16):R595–7. doi: 10.1016/s0960-9822(00)00636-9. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12(5):524–33. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Gridley T, Fishell G. Loss of notch activity in the developing central nervous system leads to increased cell death. Dev Neurosci. 2006;28(1–2):49–57. doi: 10.1159/000090752. [DOI] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Raftopoulou M, Nery S, Huang Y, Gridley T, Fishell G. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132(19):4247–58. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- Matsushita F, Kameyama T, Marunouchi T. NZF-2b is a novel predominant form of mouse NZF-2/MyT1, expressed in differentiated neurons especially at higher levels in newly generated ones. Mech Dev. 2002;118(1–2):209–13. doi: 10.1016/s0925-4773(02)00250-2. [DOI] [PubMed] [Google Scholar]
- Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, Ballivet M, Matter JM. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132(17):3907–21. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- McCabe KL, McGuire C, Reh TA. Pea3 expression is regulated by FGF signaling in the developing retina. Dev Dyn. 2006;235(2):327–35. doi: 10.1002/dvdy.20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Dotti CG. RIPped out by presenilin-dependent gamma-secretase. Cell Signal. 2003;15(9):829–41. doi: 10.1016/s0898-6568(03)00041-x. [DOI] [PubMed] [Google Scholar]
- Mellitzer G, Bonne S, Luco RF, Van De Casteele M, Lenne-Samuel N, Collombat P, Mansouri A, Lee J, Lan M, Pipeleers D, Nielsen FC, Ferrer J, Gradwohl G, Heimberg H. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25(6):1344–52. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Inazawa J, Ishida R, Tokura K, Nakahara K, Aoki K, Kasai M. Structural analysis of the gene encoding RP58, a sequence-specific transrepressor associated with heterochromatin. Gene. 2000;242(1–2):59–64. doi: 10.1016/s0378-1119(99)00477-1. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Esler WP, Kimberly WT, Jack C, Berezovska O, Kornilova A, Hyman BT, Perrimon N, Wolfe MS. Gamma-secretase/presenilin inhibitors for Alzheimer's disease phenocopy Notch mutations in Drosophila. FASEB J. 2003;17(1):79–81. doi: 10.1096/fj.02-0394fje. [DOI] [PubMed] [Google Scholar]
- Miyawaki T, Uemura A, Dezawa M, Yu RT, Ide C, Nishikawa S, Honda Y, Tanabe Y, Tanabe T. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci. 2004;24(37):8124–34. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9(6):770–8. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101(5):499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270(5233):86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228(2):151–65. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21(2):285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Matsuhashi S, Lefcort F. Restricted neural epidermal growth factor-like like 2 (NELL2) expression during muscle and neuronal differentiation. Mech Dev. 2002;119 Supl 1:S11–9. doi: 10.1016/s0925-4773(03)00084-4. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Claes K, Todd V, Chaverra M, Lefcort F. NELL2 promotes motor and sensory neuron differentiation and stimulates mitogenesis in DRG in vivo. Dev Biol. 2004;270(2):322–35. doi: 10.1016/j.ydbio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch Activity Is Downregulated Just prior to Retinal Ganglion Cell Differentiation. Dev Neurosci. 2006;28(1–2):128–41. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–89. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R. Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci. 2006;25(25):5857–65. doi: 10.1523/JNEUROSCI.4621-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target Selectivity of Vertebrate Notch Proteins: COLLABORATION BETWEEN DISCRETE DOMAINS AND CSL-BINDING SITE ARCHITECTURE DETERMINES ACTIVATION PROBABILITY. J Biol Chem. 2006;281(8):5106–19. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Price DM, Jin Z, Rabinovitch S, Campbell SD. Ectopic expression of the Drosophila Cdk1 inhibitory kinases, Wee1 and Myt1, interferes with the second mitotic wave and disrupts pattern formation during eye development. Genetics. 2002;161(2):721–31. doi: 10.1093/genetics/161.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62(18):2036–49. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Reh TA, Cagan RL. Intrinsic and extrinsic signals in the developing vertebrate and fly eyes: viewing vertebrate and invertebrate eyes in the same light. Perspect Dev Neurobiol. 1994;2(2):183–90. [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46(8):2897–904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271(2):388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8(8):995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Satow T, Bae SK, Inoue T, Inoue C, Miyoshi G, Tomita K, Bessho Y, Hashimoto N, Kageyama R. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J Neurosci. 2001;21(4):1265–73. doi: 10.1523/JNEUROSCI.21-04-01265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1–4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276(43):40268–73. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- Simmons AD, Horton S, Abney AL, Johnson JE. Neurogenin2 expression in ventral and dorsal spinal neural tube progenitor cells is regulated by distinct enhancers. Dev Biol. 2001;229(2):327–39. doi: 10.1006/dbio.2000.9984. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131(18):4447–54. doi: 10.1242/dev.01302. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–97. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Silva AO, Ercole CE, McLoon SC. Regulation of ganglion cell production by Notch signaling during retinal development. J Neurobiol. 2003;54(3):511–24. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004(1–2):148–55. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- Tokunaga A, Kohyama J, Yoshida T, Nakao K, Sawamoto K, Okano H. Mapping spatio-temporal activation of Notch signaling during neurogenesis and gliogenesis in the developing mouse brain. J Neurochem. 2004;90(1):142–54. doi: 10.1111/j.1471-4159.2004.02470.x. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16(4):723–34. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Tsai JY, Wolfe MS, Xia W. The search for gamma-secretase and development of inhibitors. Curr Med Chem. 2002;9(11):1087–106. doi: 10.2174/0929867023370185. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Research. 2003;31(24):1–8. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kim HS, Joo Y, Choi Y, Chang KA, Park CH, Shin KY, Kim S, Cheon YH, Baik TK, Kim JH, Suh YH. Intracellular domains of amyloid precursor-like protein 2 interact with CP2 transcription factor in the nucleus and induce glycogen synthase kinase-3beta expression. Cell Death Differ. 2006 2006 April 28; doi: 10.1038/sj.cdd.4401928. advance online publication. [DOI] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to Supress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133(7):1367–78. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Goto K, Kuo CH, Kondo H, Miki N. Visinin: a novel calcium binding protein expressed in retinal cone cells. Neuron. 1990;4(3):469–76. doi: 10.1016/0896-6273(90)90059-o. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20(10):1308–20. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DAPT was added to one E4.5 chick retinal explant, while DMSO was added to the sister as control. After 6h of exposure to DAPT, propidium iodide (PI) was added and incubated for 5 min. Explant pairs were washed with cold HBSS+, and flat-mounted between glass coverslips on ice. LSCM images of DMSO control (A) and DAPT-treated explant (B) from a region between the optic nerve head and folded edge of the explant. (C) Quantification of total number of PI+ cells in a given volume reveals that there is no statistical difference between DAPT-treated explants compared to control (n=3 pairs, Student's T-test, P<0.3185).
E4.5 chick retinal explant pairs were treated with DAPT for the indicated times, washed three times, cultured for a total of 48h, and immunolabeled as wholemounts with the mitotic indicator PH3 located apically, and Islet1 (Isl1) to label ganglion cells located basally. All panels are oriented such that central-to-peripheral retina is left-to-right. Images of control explants from all ages, and DAPT-treated explants from 1h and 3h exposures are taken at the folded edge of the flattened explants as depicted: single asterisk denotes normal pattern of labeling; double-headed arrows denote the edge of the explant. No effect of DAPT exposure for 1h or 3h was observed compared to controls. However periods of 6h or longer caused an permanent decrease in PH3 immunoreactivity in a central-to-peripheral wave, concomitant with increased Isl1 immunoreactivity. Note that at 6h a clear boundary is observed between the effected region located centrally, and the apparently normal region located more peripherally (dashed line).
Transient exposure to DAPT for 6h (A) produced a consistent reduction of PH3 (green) and increased Visinin (red) immunolabeling in a central-to-peripheral pattern, such that a clear boundary (arrow) was observed between the differentiated central region (single asterisk) and apparently normal peripheral region (double asterisks), which became even more apparent with 12h exposure to DAPT (B; PH3, blue; Visinin, red). The neural retina was fully differentiated with exposures to DAPT longer than 12h (data not shown).
Affymetrix arrays were used to compare global changes gene expression between E14.5 mouse retinas treated with DAPT for 8h and controls (n=12 retinas per chip, Affymetrix Mouse Genome 430 2.0 Array). Molecular changes were ranked according to initial fluorescence intensity units (FIU, > 0.5) and examined for Notch pathway associated genes. A scatter plot depicts genes identified by these criteria, and the table contains all raw information accordingly.