Abstract
A spontaneous variant of the mouse class I major histocompatibility complex Db gene, designated Dbm28, is characterized. This mutation consists of a cluster of nucleotide substitutions in exon 3 that resembles the product of a classical gene conversion event in that the substituted nucleotides appear to be templated. However, Dbm28 is distinctive, because no single donor gene containing the nucleotide sequence of the mutation exists in the genome of the parent strain. The mutation is consistent with the expected result of an interaction of two donor genes at the target locus during a single recombination event. While no known genetic mechanism gives rise to this class of mutation, we have established that 10 percent of spontaneous class I mutations in the mouse major histocompatibility complex have this complex phenotype. This process occurs at the D locus and the K locus. The significance of this kind of genetic interaction may extend beyond the major histocompatibility complex and have importance in shaping other multigene families.
Genetic recombination mechanisms play a major role in shaping the structure of genomes during evolution. Homologous recombination generates phenotypic diversity among individuals by reshuffling sequences already present within the species. Novel alleles can be generated when existing alleles involved in a crossover exhibit diversity within their coding sequences at regions that are proximal and distal to the recombination point. In a similar manner, nonhomologous recombination events between members of a multigene family also have resulted in the formation of new structural genes that can behave as new alleles (1). We have been analyzing a mechanism that generates new alleles from existing diversity within the mouse major histocompatibility complex (MHC) class I multigene family by the exchange of short stretches of sequence between ectopic members of the gene family. The structure of human class I alleles indicates that similar genetic events are occurring in human populations (2). However, new variants occur rarely and have never been observed within human family studies. Consequently, little is known about the mechanism of genetic diversification of class I genes in humans. At this time, the mouse provides the best model to study this question in a mammalian species.
Approximately 30 class I structural variants have been characterized from spontaneous “mutations” that arose in inbred mouse colonies. Analyses of the mutant nucleotide sequences reveal that in the majority of occurrences multiple nucleotide substitutions were introduced into an acceptor gene by a donor gene. Typically, these substitutions fall in a region of overall high identity between the donor and acceptor genes (3). The variants can be classified into three categories. Type I variants, such as Kbm1, have arisen by a recombination mechanism involving the gene conversion of a particular class I locus by sequences derived from another member of the class I multigene family (4–6). The resulting variant has multiple nucleotide substitutions that are identical to the sequence of the potential donor. The type II variants have a single nucleotide substitution, which can be found in other class I genes at a homologous position. Although the possibility that individual variants within this group may have occurred by a random mutation mechanism, when considered as a group, this class of variant resembles a templated event indistinguishable from the type I variants (7–10). The more perplexing group is composed of the type III variants. The structure of these variants resembles the type I and type II categories; all of the substituted nucleotides can be found in other class I genes of the parental haplotype. However, a single putative-donor gene containing all of the clustered substitutions cannot be found in the genome of the parental strain (11).
We previously have characterized two type III variants, Kbm3 and Kbm23 (11). Both contain a cluster of five nucleotide substitutions in and adjacent to the 3′ end of exon 2 of the Kb gene. In seeking an explanation for the appearance of the unusual clusters of substitutions, it may be important to consider some specific properties of the Kb locus that set this gene apart from the other members of the class I gene family. For unknown reasons, the Kb gene undergoes recombination events more frequently than other members of the multigene family. Of the 22 characterized class I variants in H-2b haplotype mice, 18 had changes at the K locus, and only 4 contained changes at the D locus. This probably was not due to selection bias, because the frequencies of changes at the K and D loci of H-2k and H-2f haplotype mice were comparable to that observed for Db, indicating that Kb is more susceptible to these changes than other loci. Furthermore, 5 of the 18 variants contained substitutions in the region of the Kb gene affected by the bm3 and bm23 mutations. Therefore, on the basis of the unusual recombination activity of this region of the Kb gene, one might consider the possibility that the type III mutations arise as a result of sequential type I gene conversion events. We have argued that this is probably not the case, because even at the inflated frequencies observed for the Kb gene, and allowing for the possibility that silent substitutions occurred in generations preceding and following the generation of a functional variant, it is highly improbable that two independent type III variants would have been observed among the 30 characterized class I variants (11).
We now describe a third example of a type III variant, Dbm28. Dbm28 is significant, because in this case the substitutions have occurred at the Db locus and in a region of the gene that has not been previously indicated as a hot spot for recombinatory events. The identification of Dbm28 as a type III variant argues that this class of variant does not arise by sequential type I mutations that happen at an unexpectedly high frequency due to the unique properties of the Kb gene or because they have occurred in a general recombinatory hotspot of a class I gene. This generalization of the mechanism leading to type III variants to more than one class I locus strengthens the hypothesis that a novel recombination mechanism may involve the interaction of more than two homologous genes. Such a mechanism could have profound influence on the evolution of multigene families.
MATERIALS AND METHODS
Mice.
C57BL/6By-H-2bm28 mice (bm28) were bred in the immunogenetics mouse colony at Northwestern University. This variant arose in 1981 in D. W. Bailey’s mouse colony of C57BL/6By mice at the Jackson Laboratory (12). The mutation, detected by skin graft incompatibility, occurred in a pedigree in which the parents were screened using skin grafts and found to be fully histocompatible with other screened members of the strain. The mutation, designated A-2e and later renamed bm28, has been maintained on the C57BL/6By background by breeding sibs.
Generation of cDNA and Cloning.
RNA was isolated from the livers of bm28 mice using TRIzol reagent (GIBCO/BRL). We generated single-stranded cDNA using cDNA Synthesis SystemPlus (Amersham) from 20 μg of total RNA, then amplified the full-length Dbm28 cDNA using PCR with D locus-specific primers, as previously described (13). By engineering BamHI sites into the D locus-specific primers, we could clone the product into the unique BamHI site of the pUC18 vector. Clones containing Dbm28 cDNA insert hybridized to an oligonucleotide probe specific for the Db exon 4 sequences (Ld-191 TCACCCCAGATCTAAAGGTG). The procedure was repeated independently to ensure that PCR artifacts would be detected. The observed mutations were common to the independently isolated cDNA.
Sequence Analysis of Cloned Inserts.
The DNA insert of interest was sequenced using a Sanger-based fluorescent dideoxynucleotide method with an automated DNA sequencing apparatus (model 377, Applied Biosystems). Approximately 3.2 pmol of oligonucleotide primer was used to sequence 1 μg of plasmid containing insert. The sequence data was analyzed using the Wisconsin Package, Version 8.1 (Genetics Computer Group, Madison, WI).
Cosmid Clones and Genomic DNA.
The cosmid library containing K, Q, D, and T region genes was a gift from G. L. Waneck (Harvard Medical School, Boston). As previously described (14), the overlapping cosmids completely represent the H-2b class Ia and Ib genes that reside in the K, D/Q, or T regions. We isolated splenic genomic DNA from C57BL/6ByJ, using a method described previously (15). From the bm28 mice, we used a variation on this protocol to isolate liver DNA. The liver was finely minced, then digested overnight in buffer containing 10 mM Tris·HCl at pH 7.4, 100 mM EDTA, 250 μg/ml proteinase K/0, and .5% SDS.
Southern Blots.
For the Southern blots of the cosmid library, we digested approximately 2 μg of cosmid DNA with BamHI endonuclease. The size-separated fragments were transferred onto Magna nylon membranes (Micron Separations, Westboro, MA), and hybridized to the 32P-labeled oligonucleotide probe. The membranes were prehybridized in 50 ml of 5× standard saline phosphate EDTA (SSPE) (1× SSPE is 150 mM NaCl/10 mM NaH2PO4·H2O/1 mM EDTA), 1% SDS, and 100 μg/ml denatured salmon sperm DNA (Sigma). The oligonucleotide probe was end-labeled with polynucleotide kinase, then added to the prehybridizing membranes. The hybridizing and washing conditions were at stringencies that allowed detection of a plasmid containing Dbm28 cDNA (pUCDbm28), but not a plasmid containing a genomic clone of the Db gene. The temperatures that we used to hybridize the DNA to the probe and to wash the membranes were estimated by using the formula Tw (°C) = 2 × [(A + T) content] + 4 × [(G + C) content]. Genomic Southern blotting was performed using previously described methods (7). We used BamHI, BglII, or HindIII to digest 40 μg of B6 or bm28 genomic DNA. As for the cosmid Southern blots, we transferred the size-separated fragments onto Magna nylon membranes, then hybridized to a 32P-labeled oligonucleotide probe. The temperatures that we used to hybridize the digested DNA to the probe and to wash the filters were estimated by using the formula Tw (°C) = 2 × [(A + T) content] + 4 × [(G + C) content] − 5. We modified the wash conditions from the previous procedure, which resulted in better resolution of the hybridizing bands. The first wash solution was 3× SSC/10 mM sodium phosphate, pH 7.0/10× Denhardt’s solution/5% SDS. One times SSC is 150 mM NaCl/15 mM sodium citrate; and 1× Denhardt’s solution is 0.02% Ficoll/0.02% BSA/0.02% polyvinylpyrrolidone. We incubated the filters at 42°C, then at Tw, for 1 h at each temperature. The filters were transferred to the second wash solution (1× SSC/1% SDS) and incubated at Tw for 1 h. The oligonucleotide probes used in this study were Dbm28-97/99–1 (5′-GTGGATGTATGGCTGTGACCT-3′), Dbm28-97/99–2 (5′-ACACTCCAGTGGATGTATGGC-3′), and Dbm28-103 (5′-GCTGTGACCTGGGGTCGGA-3′).
RESULTS
Origin and Characterization of bm28.
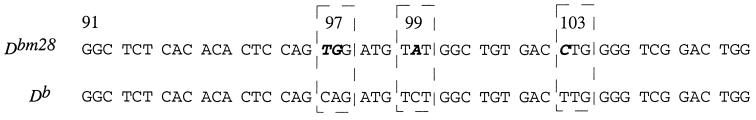
The bm28 mouse was identified by skin graft rejection using the C57BL/6By mouse strain and mapped to the D locus using traditional complementation analysis (12). Using D locus-specific primers in a PCR, we cloned the Dbm28 transcript by amplifying reverse-transcribed cDNA and sequenced four independent clones. Four differences in the nucleotide sequence between the parent D and the mutant Dbm28 gene were identified (Fig. 1). The mutation cluster affects codons 97, 99, and 103. The substitutions at 97 (Q to W) and 99 (S to Y) are productive, and the substitution at 103 is silent. These results were confirmed in completely independent experiments, including PCR amplifications. The presence of the identified mutations in the bm28 genome was confirmed by genomic Southern blot analysis, as discussed below. The substituted amino acid positions are located in the peptide binding cleft of the class I molecule. These positions are consistent with the alloantigenicity of the variant D molecule detected in the skin graft screen.
Figure 1.
Nucleotide sequence comparison of the Dbm28 mutation with the parental Db gene reveals a clustered nucleotide substitution. The region of the gene that contains the mutation cluster is depicted; the rest of the cDNA sequence otherwise matches the known coding sequence of the Db gene (16). The mutations are productive at codons 97 and 99, which result in amino acid changes Q97W and S99Y. The mutation at codon 103 is silent.
Absence of a Single Donor Gene.
Initial comparisons of the Dbm28 mutation cluster with the previously reported sequences of the H-2b classical and nonclassical genes immediately ruled out these genes as potential gene conversion donors. Notably, the K and K1 loci were not the donor genes (17), demonstrating that unlike the previously characterized D mutants (8, 9), the bm28 mutation is the first conversion of the D gene that did not use the K gene as a donor. Although there are genes that have homology to the cluster at 97 and 99 or for the substitution at 103, no single gene has the entire mutation cluster.
Because the donor genes for all mutants characterized to date have been located in the K, D, Q, or T regions (6), we pursued the possibility that an uncharacterized donor gene exists for the bm28 mutation cluster within the Q or T regions of the class I gene family. We designed oligonucleotide probes specific for either the 97/99 (probe Dbm28-97/99–1) or 103 (probe Dbm28-103) mutations and screened a library of overlapping cosmids spanning the D and Q regions or the T region of the genome. Hybridization and washes under stringent conditions allowed detection of a plasmid containing a Dbm28 cDNA clone, but not of a plasmid containing a Db genomic clone.
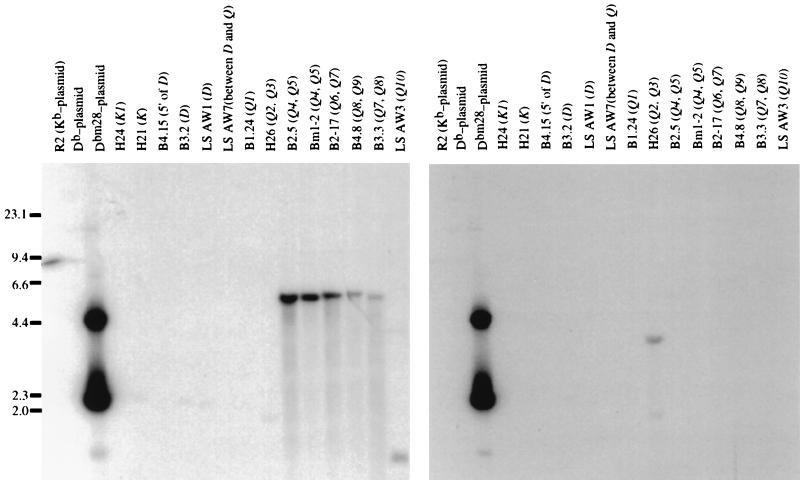
Screening of the D and Q region cosmids revealed multiple genes that have either the nucleotide sequence at positions 97 and 99, comprising the left half of the mutation cluster, or the nucleotide at 103, comprising the right half of the cluster (Fig. 2). The probe specific for the left half hybridized to ≈5.5-kb BamHI fragments from cosmids representing the Q4 through Q9 genes. From the known restriction map of the cosmid library, the size of these bands is consistent with the expected BamHI fragment of Q5, Q6, Q7, Q8, and Q9 that contains the region of interest. The positive ≈1.0-kb BamHI fragment of the cosmid containing Q10 is consistent with the known BamHI restriction sites in the Q10 gene. Using the probe specific for the nucleotide substitution at codon 103, we identified a ≈3.8-kb BamHI fragment from the cosmid containing Q2 and Q3. This fragment’s size implicates Q2 as a potential donor gene. Thus, our analysis of the Q region class I genes demonstrates that several genes hybridize to probes defining either the left half or right half of the bm28 mutation cluster, but no single gene hybridizes to both probes.
Figure 2.
Sequences of Q region genes match either the 5′ (left half) or 3′ (right half) region of the bm28 mutation cluster, but no single gene in the Q region matches the entire mutation. We hybridized the BamHI-digested cosmids representing the H-2b K and D/Q region genes with oligonucleotide probes that were specific for the left (Dbm28-97/99–1) or right (Dbm28-103) half of the mutation cluster. Multiple genes from the Q region contain nucleotides in common with bm28 codons 97 and 99 (Left), and Q2 contains the single nucleotide match with bm28 codon 103 (Right).
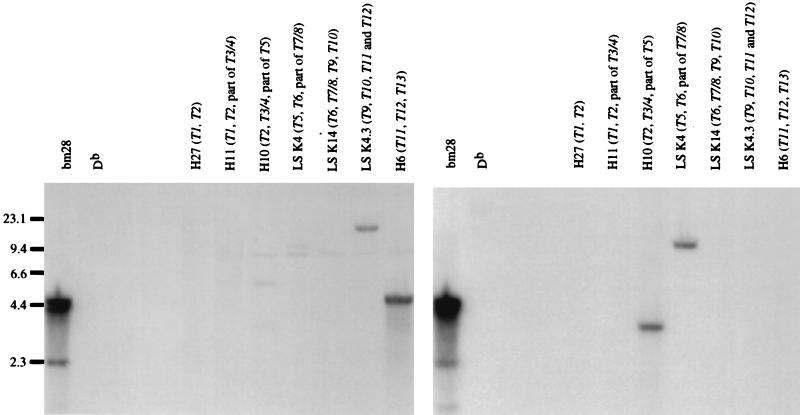
Similarly, when we screened cosmids spanning the T region, we identified genes that were positive for either the left half or right half of the mutation cluster, but no gene in this region contained both (Fig. 3). The left-half probe hybridized to fragments from cosmids that span the T9 to T12 genes. T12 is partially represented in cosmid LS K 4.3 and completely represented in cosmid H6; the positive ≈4.8-kb BamHI fragment of the H6 digested DNA suggests that T12 contains the left half of the cluster. The right-half probe hybridized to fragments from cosmids that completely span the T3 to T7 genes and include the 3′ portion of T2 and the 5′ portion of T8. In particular, T5 is partially represented in cosmid H10 and completely represented in cosmid LS K 4. The right-half probe hybridized to a ≈13.9-kb BamHI fragment in the LS K 4 digested cosmid, consistent with the expected size of a BamHI fragment that contains the T5 gene.
Figure 3.
Sequences of T region genes match either the left half or right half of the bm28 mutation cluster, but no single gene in the T region matches the entire mutation. We hybridized the BamHI-digested cosmids representing the H-2b T region genes with oligonucleotide probes that were specific for the left (Dbm28-97/99–1) or right (Dbm28-103) half of the mutation cluster. T12 contains nucleotides that are homologous to the left half of the mutation cluster (Left), and T5 has the right half of the mutation cluster (Right).
From the cosmid hybridization analysis, we predicted that Q5, Q6, Q7, Q8, Q9, Q10, and T12 had the sequences at codons 97 and 99, and that Q2 and T5 had the sequence at 103. We verified this prediction by analysis of the published nucleotide sequences for most of these genes (18–22) (Fig. 4). The sequence for T12 was not available. To verify that this gene contained the left, but not right, half of the mutation cluster, we subcloned the H6 cosmid BamHI fragment that hybridized to the left-half probe and sequenced across the region of codons 97, 99, and 103. As shown in Fig. 4, T12 contained the predicted sequence targeted by the probe, but it did not contain the entire mutation cluster. There are a number of candidate donor genes for the left half of the mutation cluster, most with significant stretches of nucleotide homology. Of the candidate donor genes for the right half of the cluster, T5 has the longest stretch of nucleotide homology. When analyzing the MHC class I gene sequences in the databank and in published sources, we noticed that Q1 also had the homologous nucleotide at codon 103 (ref. 17). Q1 is an unlikely donor gene for this event, because, within the mutation cluster, it has an additional mismatch with Dbm28. From the sequence and cosmid analyses, we conclude that a donor gene containing homologous sequence to bm28 at codons 97, 99, and 103 was not present in the K, Q, or T region genes.
Figure 4.
The bm28 mutation cluster’s homology to Q and T region genes demonstrates the nonrandom nature of this mutation event. Using the data from the Southern blot analysis of the cosmid library with the left-half or right-half probes to the mutation cluster, we analyzed the known sequence of the genes. Because there is no published sequence for T12, we isolated the hybridizing BamHI fragment from H6, cloned it, and sequenced across the 5′ region of exon 3. The figure shows the partial nucleotide sequence for the region of interest from each gene, with the candidate donor genes’ nucleotide differences emphasized in lowercase bold letters.
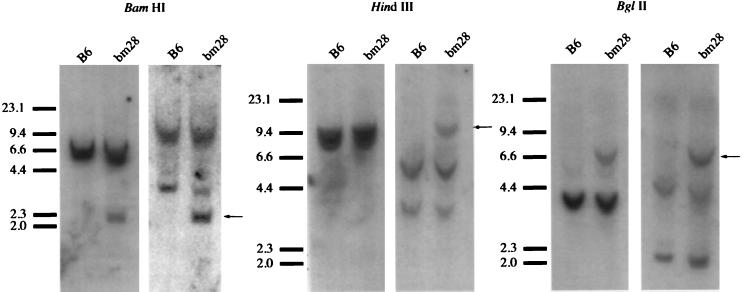
Because there remained a possibility that a donor gene existed outside of the region represented by the cosmid library, we extended our search to the entire C57BL/6 genome by performing a series of genomic Southern blots with DNA from C57BL/6 and bm28 mice. The DNA was fragmented with BamHI, HindIII, or BglII. We hybridized the DNA with oligonucleotide probes specific for either the 97/99 or 103 mutations under stringent conditions that allowed detection of the fragment containing D gene in the bm28 genomic DNA, but not the homologous D gene fragment in the C57BL/6 DNA (Fig. 5). Under these conditions, a restriction fragment that contains a possible donor gene for the bm28 mutation will hybridize to both oligonucleotide probes.
Figure 5.
There is no single gene in the parental strain (C57BL/6) genome that could have given rise to the bm28 mutation. We probed the B6 genome for genes that might contain both halves of the bm28 mutation cluster. Genomic DNA from bm28 mice and B6 mice was digested with one of three endonucleases, size-separated, then hybridized to probe Dbm28-97/99–2 (left blot in each pair) or to probe Dbm28-103 (right blot in each pair). Any fragment that contains a gene with the entire mutation cluster would have a consistent size when hybridized to either the left- or right-half probe; however, the only fragment with this property is the fragment containing the Dbm28 gene (marked by arrows).
In the genomic DNA digested with BamHI, HindIII, or BglII, multiple bands hybridized to either probe, consistent with the Southern blot analysis of the cosmid library and the nucleotide sequence data where we observed multiple genes containing either half of the mutation. In addition, both probes were able to detect the fragment harboring the Dbm28 gene, but not the wild-type Db gene, confirming the presence of the detected nucleotide changes in the variant (Fig. 5). For example, in the BamHI-digested bm28 DNA, both probes detected the ≈2.1-kb fragment with the Dbm28 locus. In addition, the right-hand probe hybridized to a >14-kb and a <4-kb fragment, but the left-half probe hybridized to only a <6.0-kb fragment. However, no comigrating fragments hybridized to both probes, demonstrating that no single donor gene in the parental genome contains the entire sequence cluster at codons 97, 99, and 103. Similar hybridization patterns were observed with HindIII- and BglII-digested genomic DNA, strengthening the evidence against the existence of a single donor gene in the genome.
Mismatch Repair Errors.
One explanation for the appearance of type III nucleotide substitution profiles would involve interaction between two genes, a single donor and a single acceptor locus. In the gene conversion process, random errors during repair of heteroduplex intermediates might result in “mutation clusters” that resemble a recombination product between two donor genes. Due to the high degree of diversity within this multigene family, one would expect that random mutations caused by polymerase-based errors of replication occasionally would yield a stretch of substitutions that are identical in sequence to homologous regions of other genes within the multigene family, as found in all three type III variants characterized to date. However, it is more likely that random nucleotide misincorporations would result in sequences that are distinct from any of those in the class I multigene family. To estimate the probability that a random substitution would fortuitously match known class I sequences within the parental haplotype, we tabulated all the bases that occur in exons 2 and 3 (the sites for all known class I gene conversion events) in two panels of published mouse class I genes that comprise 22 K and D alleles and 10 Q region genes (6, 19). A random mutation would match a nucleotide already present in this panel with a frequency of 0.217 (21.7% of the possible nucleotides are represented in the panel of sequences). The probability that this would happen on three independent occasions, as represented by bm3, bm23, and bm28, would be approximately 0.01. Furthermore, this is a conservative estimate, because only a few genes within the parental haplotype seem to participate in the gene conversion process. Another fact that argues against the occurrence of a random error is that, to explain the Kbm3 mutation, multiple errors would have had to be introduced, because there does not appear to be a single donor gene candidate that is only one nucleotide different from the variant. Therefore, it is highly unlikely that type III substitution clusters are generated by random base substitutions during the course of gene conversion.
DISCUSSION
We have demonstrated that Kbm3, Kbm23, and Dbm28 are three independent examples of substituted nucleotide clusters in mouse class I genes that do not fit the profile of simple gene conversion recombinants. Each of these variants contains multiple nucleotide substitutions that individually can be found in other class I genes within the parental MHC haplotype. However, no single parental gene has been detected that contains the entire substituted sequence. It may be significant that genes known to have served as donor genes in the traditional type I gene conversion events (D, Q4, Q10, and T5) contain partial sequences that match either the left or right halves of the substitution clusters defined in the type III variants. These findings are consistent with the hypothesis that this class of variant is derived from a previously undescribed complex gene conversion event that involves two donor genes. This complex recombinatory event is not particularly rare, because 10% of the class I variants analyzed so far have had this structure.
The documentation of a type III substitution at the D locus extends our observations of this kind of genetic interaction, which previously were limited to a single region within Kb. Kb has been observed to recombine at a frequency approximately 5 to 10 times higher than other class I genes and to contain regions that are relatively frequent targets for gene conversion. The Db locus has no history of a high recombination rate, nor are the changes in the bm28 variant in a region of the gene previously associated with multiple recombination events. This suggests that the genetic events that generate the type III MHC variants are not specific to the Kb gene, but, in fact, may be a general mechanism that permits the flow of genetic information among members of multigene families. MHC class I genes may simply provide a unique opportunity to visualize these recombination events, because they result in functional changes in the antigen-presenting properties of the encoded molecules, which can be selected by an immunological reaction. Visualization of variants in other gene families would require molecular assays capable of screening thousands of individuals or an analysis that can take advantage of the individual nature of germ-line cells such as spermatozoa (23). Approaches of this kind also would permit similar analyses to be conducted in other species, such as humans, where screening large numbers of genetically typed animals is not possible.
The central unaddressed issue is the nature of the genetic process that gives rise to the type III variants. No precedent provides a satisfactory model for how these genetic interactions might be taking place. Simultaneous pairing between three loci during a single recombination event has not been described. A model involving two sequential conversion events by two donor genes is appealing in its simplicity. However, its weakness is that the occurrence of consecutive independent gene conversion events is highly improbable (even allowing for multiple generations that separate variants from their time of analysis) based both on the low frequency of independent events (<10−3 per event) and the fact that the required two events, in each case, must affect the same small stretch of nucleotides. To account for this latter observation, we would have to propose either of two additional possible mechanisms for this model. First, an unknown mechanism may be able to target a region of the gene that would result in the contiguous clusters of substituted nucleotides characteristic of these variants. An alternative to a targeted mechanism is an inductive mechanism where a single conversion event induces a second conversion within the same region. To test such elaborate variations of a sequential model, characterization of the actual enzymatic machinery and the genetic substrates that are involved in the event is required.
Another attractive explanation is that the type III substitution clusters arise as a result of errors of replication upon heteroduplex resolution during gene conversion. However, random replication errors would not be expected to result in nucleotide changes that are already present within the class I multigene family, especially if one limits the candidate donor genes to the few members that have been consistently implicated in previously described gene conversion events. With the description of Dbm28, the probability that random changes account for our inability to identify donor genes can be rejected by statistical considerations. This analysis leaves us with the conclusion that the nucleotide substitutions are not randomly determined.
There are two plausible mechanisms for the appearance of nonrandom nucleotide substitutions. The first is that the structure of heteroduplexed nucleotide stretches being resolved can lead to specific errors during replication. Such errors could recur periodically throughout the multigene family, such that other genes (putative donor genes) may have undergone identical changes independently during the course of the evolution of the class I multigene family (24). One weakness of this model is that the nucleotides attributed to replication error would have to be present within the members of the parental haplotype, which would be unlikely unless there is a very strong bias for the occurrence of particular mutations. For example, the nucleotide substitution that gives rise to the silent base change at position 103 in the bm28 mutation is present in several Q and T region genes, but is not present in any of the K and D alleles sequenced to date (6, 19). If this were a sequence-directed replication error of appreciable frequency, one would expect to see this polymorphism among the K and D alleles.
The second mechanism by which nonrandom nucleotide substitutions can appear is that more than one donor gene templated the substitutions. This model implies the interaction of at least three genes in the gene conversion event. We previously have discussed one model that could give rise to type III substitutions, and we proposed that an interaction between two donor genes occurs during an intermediate step before the actual conversion of the acceptor locus (11). This is currently our favored hypothesis, because it explains all the observations of the spontaneous mutations. The current challenge is to develop systems that improve the current detection method to test the tenets and predictions of this model, and we are pursuing several strategies to increase the level and efficiency of detecting spontaneous mutations.
Our characterization of the bm28 mutation solidifies the identification of an unusual recombination mechanism that is occurring in mice. Such a mechanism, where sequences from more than one gene can be introduced into another during a single recombination event, has not been previously described in vivo to our knowledge. This finding may not be limited to the MHC multigene family. The significance of this mechanism is that by mediating the flow of genetic information among related members of gene families, the mechanism could have profound implications for how they evolve.
Acknowledgments
We thank R. J. Pogulis, A. N. Vallejo, S. T. Kuhns, and M. D. Tallquist for critical review of this manuscript and for helpful discussions, Rudy D. Hanson for technical assistance, and Terri Felmlee for secretarial support. This work was supported by National Institutes of Health Grants AI16919 and AI22420.
Footnotes
References
- 1.Sun Y H, Goodenow R S, Hood L H. J Exp Med. 1985;162:1588–1602. doi: 10.1084/jem.162.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parham P, Lawlor D A, Lomen C E, Ennis P D. J Immunol. 1989;142:3937–3950. [PubMed] [Google Scholar]
- 3.Nathenson S G, Geliebter J, Pfaffenbach G M, Zeff R A. Ann Rev Immunol. 1986;4:471–502. doi: 10.1146/annurev.iy.04.040186.002351. [DOI] [PubMed] [Google Scholar]
- 4.Mellor A L, Weiss E H, Kress M, Jay G, Flavell R A. Nature (London) 1984;306:792–795. [Google Scholar]
- 5.Pease L R, Schulze D H, Pfaffenbach G M, Nathenson S G. Proc Natl Acad Sci USA. 1983;80:242–246. doi: 10.1073/pnas.80.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pease L R, Horton R M, Pullen J K, Cai Z. Crit Rev Immunol. 1991;11:1–32. [PubMed] [Google Scholar]
- 7.Geliebter J, Zeff R A, Schulze D H, Pease L R, Weiss E H, Mellor A L, Flavell R A, Nathenson S G. Mol Cell Biol. 1986;6:645–652. doi: 10.1128/mcb.6.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmi S, Geliebter J, Zeff R A, Melvold R W, Nathenson S G. J Exp Med. 1988;168:2319–2335. doi: 10.1084/jem.168.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrand W H, Horton R M, Pease L R, Martinko J M. Mol Immunol. 1992;29:61–69. doi: 10.1016/0161-5890(92)90157-s. [DOI] [PubMed] [Google Scholar]
- 10.Horton R M, Hildebrand W H, Martinko J M, Pease L R. J Immunol. 1990;145:1782–1787. [PubMed] [Google Scholar]
- 11.Pease L R, Horton R M, Pullen J K, Yun T J. Mol Cell Biol. 1993;13:4374–4381. doi: 10.1128/mcb.13.7.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melvold R W. Mouse News Letter. 1989;84:58. [Google Scholar]
- 13.Cai Z, Pullen J K, Horton R M, Pease L R. Methods Enzymol. 1992;216:100–108. doi: 10.1016/0076-6879(92)16012-9. [DOI] [PubMed] [Google Scholar]
- 14.Weiss E H, Golden L, Fahrner K, Mellor A L, Devlin J J, Bullman H, Tiddens H, Bud H, Flavell R A. Nature (London) 1984;310:650–655. doi: 10.1038/310650a0. [DOI] [PubMed] [Google Scholar]
- 15.Pease L R, Nathenson S G, Leinwand L A. Nature (London) 1982;298:382–385. doi: 10.1038/298382a0. [DOI] [PubMed] [Google Scholar]
- 16.Duran L W, Horton R M, Birschbach C W, Chang-Miller A, Pease L R. J Immunol. 1989;142:288–296. [PubMed] [Google Scholar]
- 17.Weiss E H, Golden L, Zakut R, Mellor A, Fahrner K, Kvist S, Flavell R A. EMBO J. 1983;2:453–462. doi: 10.1002/j.1460-2075.1983.tb01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geliebter J, Nathenson S G. Mol Cell Biol. 1988;8:4342–4352. doi: 10.1128/mcb.8.10.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhner M K, Goodenow R S. Immunogenetics. 1989;30:458–464. doi: 10.1007/BF02421178. [DOI] [PubMed] [Google Scholar]
- 20.Cullen M K, Lapierre L A, Kesari K V, Geliebter J. J Exp Med. 1993;177:1803–1807. doi: 10.1084/jem.177.6.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin J J, Weiss E H, Paulson M, Flavell R A. EMBO J. 1985;4:3203–3207. doi: 10.1002/j.1460-2075.1985.tb04066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson P J, Bevec D, Mellor A L, Weiss E H. Immunogenetics. 1988;27:79–86. doi: 10.1007/BF00351079. [DOI] [PubMed] [Google Scholar]
- 23.Murti J R, Bumbulis M, Schimenti J C. Mol Cell Biol. 1992;12:2545–2552. doi: 10.1128/mcb.12.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein J. Transplantation. 1984;38:327–329. doi: 10.1097/00007890-198410000-00002. [DOI] [PubMed] [Google Scholar]