Abstract
The ELL gene was first identified by its involvement with MLL in the translocation (11;19)(q23;p13.1) in acute myeloid leukemia. To date, nine other MLL partner genes have been cloned, but their precise functions have yet to be determined. To characterize the functions of ELL further, we have cloned the murine homologue of ELL and have found that the gene is highly conserved at the nucleotide and amino acid level. The open reading frame of the murine homologue contains 602 aa, slightly smaller than the 621 aa in the human gene. With Northern blot analysis, a 3.4-kb transcript is detected in all tissues examined with greatest levels of expression in the liver. Unlike human ELL, only a single transcript can be detected with either murine coding sequence or 3′ untranslated region probes. To examine the spatial and temporal pattern of expression in murine development, in situ hybridization studies were performed with sense and antisense riboprobes from the 3′ untranslated region of murine Ell. Ell is expressed diffusely by embryonic day 7.5 (E7.5). In addition, high levels of expression can be detected in maternally derived decidual tissue. At E14.5, Ell is expressed diffusely throughout the embryo. However by E16.5, specific expression in the liver and gastrointestinal tract becomes prominent and remains so in both neonates and adults. To determine the subcellular localization of ELL, we developed a polyclonal antiserum to ELL that was used for immunofluorescence studies in COS-7, HeLa, NIH 3T3, and A7r5 cells. The ELL protein was localized to the nucleus but excluded from nucleoli in all cell lines examined. Recently, the gene product of ELL was found to function as an RNA polymerase II elongation factor, an activity that is consistent with our immunofluorescence data. Thus, these studies extend our understanding of the normal functions of ELL and provide additional insight into its aberrant function when fused to MLL in acute myeloid leukemia.
We first identified the ELL gene as a partner gene of MLL in the translocation (11;19)(q23;p13.1), a recurring cytogenetic abnormality in de novo and therapy-related acute myeloid leukemia (1). Previously, we found that the MLL gene is involved in over 20 different cytogenetic aberrations that affect chromosome band 11q23 (2). The majority of these events are reciprocal translocations, and less commonly, some are insertions or inversions, which result in the juxtaposition of the MLL gene with sequences located on other chromosomes. The critical feature of these chromosomal rearrangements is the generation of an in frame chimeric fusion transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome.
Nine other genes at 11q23 partner chromosomal breakpoints have been cloned. These include AF-4 in the t(4;11)(q21;q23) (3), ENL in the t(11;19)(q23;p13.3) (4), AF-9 in the t(9;11)(p22;q23) (5), AF-6 in the t(6;11)(q27;q23) (6), AF-1p in the t(1;11)(p32;q23) (7), AF-X in the t(X;11)(q13;q23) (8), AF-1q in the t(1;11)(q21;23) (9), AF-10 in the t(10;11)(p13;q23) (10), and AF-17 in the t(11;17)(q21;q23) (11). Although AF-10 and AF-17 appear to be members of a new gene family and AF-9 and ENL share homology at their C termini, no consistent homologies have been identified among the partner gene sequences that would explain how so many different genes can fuse to MLL and all be leukemogenic. The functions of these other fusion partner genes have yet to be determined. Recently, Corral et al. (12) used homologous recombination to generate an Mll–AF-9 fusion gene in murine embryonic stem cells (12). The chimeric mice generated with these embryonic stem cells developed acute myeloid leukemia, confirming that the partner genes are critical to leukemogenesis.
As a result of the t(11;19)(q23;p13.1), an MLL–ELL chimeric fusion gene is created. This transcript contains 5′ MLL sequences including the AT hooks, a methyltransferase domain, a proline-rich region, and a repression domain. The zinc fingers and the remainder of MLL, including the region with the highest homology to the Drosophila trithorax gene, are not part of the critical fusion transcript.§ In the t(11;19)(q23;p13.1), all but 124 of the most 5′ nucleotides of ELL are fused to MLL to generate the chimeric transcript. Variable amounts of the 3′ sequences of translocation partner genes fuse to MLL in other 11q23 translocations.
At the time that we completed the sequencing of ELL, database searches did not reveal significant homologies to other known genes. The only conserved motif was a highly basic, lysine-rich motif of ELL that is homologous to similar regions of several proteins, including the DNA binding domain of poly(ADP·ribose) polymerase. We named the gene ELL for eleven–nineteen lysine-rich leukemia gene. ELL contains a predicted open reading frame of 621 aa; 576 aa of 3′ ELL fuse to 5′ MLL sequences as a result of the translocation. ELL is not homologous to other MLL partner genes. Recently, ELL was found to function as an RNA polymerase II transcription elongation factor (13). It serves to increase the catalytic rate of RNA polymerase II transcription by suppressing transient pausing by the polymerase along the DNA template. The elongin gene has previously been identified to have a similar function (14). Elongin has been reported to be regulated by the VHL (for von Hippel–Lindau) tumor suppressor gene (15). The aberrant functions of ELL when fused to MLL remain to be determined.
In this study, we report the cloning of the murine homologue of the ELL gene. Sequence comparisons reveal a high degree of evolutionary conservation, suggesting sequence constraints in its functional motifs. 5′ rapid amplification of cDNA ends (RACE) on the murine sequence supports the assignment of the initiating methionine that we identified in human ELL. To examine the spatial and temporal pattern of expression of Ell in murine development, we have performed in situ hybridization studies with sense and antisense riboprobes derived from the 3′ untranslated region (UTR) of Ell. We have identified a diffuse pattern of Ell expression early in embryonic development, but in later stage embryos and adults, we observed a more specific pattern of hybridization with higher Ell expression in the liver and gastrointestinal tract. In addition, to determine its subcellular localization, we have generated a polyclonal antiserum to ELL that was used for immunofluorescence studies. We identified a pattern of diffuse nucleoplasmic expression with exclusion of nucleoli that is similar to the localization previously observed for RNA polymerase II (16), supporting the functional data that ELL acts as an RNA polymerase II transcription elongation factor. Because ELL is the first MLL partner gene with a known function, these experiments, in addition to characterizing ELL further, provide additional insights into the potential mechanisms of 11q23 leukemias.
MATERIALS AND METHODS
Screening of cDNA Libraries.
A cDNA library prepared in the lambda gt11 vector from murine myeloma J558L cells (Stratagene) was screened with an oligolabeled 1.8-kb fragment that spanned the open reading frame of ELL. Five positive clones were isolated and plaque was purified. Phage DNA was digested with BsiWI and the inserts were subcloned into pSL1190.
Nucleotide Sequencing.
cDNA clones were sequenced on both strands using dye terminators and AmpliTaq DNA polymerase (Perkin–Elmer) for cycle sequencing. The reactions were run on an Applied Biosystems model 377 sequencing apparatus. Chromatograms were imported into the sequencher program (Gene Codes, Ann Arbor, MI) for review and for assembly of contigs. Database searches were conducted with the blastn and blastp programs through GenBank and with the fasta and tfasta programs through the GCG Wisconsin sequence analysis package. Evolutionary comparisons were conducted with the new diverge program from GCG.
5′ RACE.
Mouse liver cDNA (marathon-ready cDNA; CLONTECH) was obtained as a template for 5′ RACE PCR. A nested PCR strategy was used. The primers for the first round of PCR were an antisense 28-mer oligonucleotide extending from nucleotide 386 to nucleotide 359 of Ell and an anchor primer. A 1-μl aliquot of this reaction was used as the template for the second round of nested PCR using an internal anchor primer and an Ell primer extending from nucleotide 360 to nucleotide 342. The products from the second round of amplification were gel-purified and subcloned by standard techniques. Ten individual clones were isolated and sequenced.
Northern Blot Analysis.
A multiple-tissue Northern blot was hybridized and washed according to the manufacturer’s suggestions (CLONTECH). The blot was then hybridized with an actin probe to determine the amount of RNA loaded in each lane. To measure band intensity, the autoradiographs were scanned by a Molecular Dynamics densitometer.
In Situ Hybridization.
Sections of murine embryonic and adult tissues were mounted for in situ hybridization. A 535-bp fragment of the murine Ell 3′ UTR spanning nucleotides 1797–2332 was amplified by PCR and subcloned into the pNOTA vector (5 Prime → 3 Prime). Sense and antisense riboprobes were labeled with [35S]dUTP (Amersham) using T7 RNA polymerase (Promega). Tissues were fixed in 4% paraformaldehyde/PBS and embedded in paraffin. The sections were rehydrated and treated with 20 mg/ml proteinase K and 0.1 M triethanloamide·HCl (pH 8). Pretreated slides were hybridized overnight at 60°C in 50% formamide, 20 mM Tris·HCl (pH 8), 0.3 mM NaCl, 5 mM EDTA, 10% dextran sulfate, 0.02% BSA, 0.5 mg/ml tRNA, 10 mM DTT, 2.5 μM 5′-α-thio-ATP, and 35S-labeled riboprobe [140,000 cpm/μl (90 μl per slide)]. After hybridization, slides were washed with 50% formamide, 2× standard saline citrate (SSC), and 20 mM DTT for 60 min at 65°C (high-stringency wash). Slides were then washed with STE (4× SSC/20 mM Tris·HCl, pH 7.6/1 mM EDTA) for 10 min twice at 37°C, followed by RNase A treatment (10 mg/ml in STE) for 30 min at 37°C. This high-stringency wash was repeated, and the slides were then washed in 2× SSC at 37°C for 10 min. Sections were processed for emulsion autoradiography, poststained with Hoechst 33258, and visualized by epifluorescence and darkfield microscopy on a Zeiss Axiophot microscope.
Generation of Polyclonal Antiserum.
The C-terminal coding sequence of human ELL from amino acid 250 to amino acid 621 was subcloned into the SmaI and SalI sites of the bacterial expression vector pGEX-4T3 (Pharmacia). A glutathione S-transferase (GST)–ELL fusion protein was induced at 1.25 mM isopropyl β-d-thiogalactoside (IPTG) and subsequently purified using the Bulk GST Purification Module (Pharmacia). Polyclonal anti-ELL antisera were obtained from rabbits immunized with the GST–ELL fusion protein (Pocono Rabbit Farm, Canadensis, PA). By Western blot analysis, the polyclonal anti-ELL antisera hybridized specifically to the GST–ELL fusion protein but not to GST alone or to an unrelated GST–LIM protein. In addition, rabbit reticulocyte lysates were used to perform coupled in vitro transcription and translation reactions with the PCDNA3 vector alone, PCDNA3 containing full-length ELL, or PCDNA3 containing an unrelated LIM protein. The products of these reactions were blotted with the preimmune and polyclonal anti-ELL antisera, and a specific band of the appropriate size was identified by the polyclonal anti-ELL antisera that was not present in the control lanes or with the preimmune sera (data not shown).
Immunofluorescence.
The cell lines used in these studies were as follows: A7r5 (rat), NIH 3T3 (mouse), COS-7 (monkey), and HeLa (human). A7r5 is a smooth muscle cell line derived from rat embryonic aorta. The cells were grown on coverslips in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (GIBCO/BRL). The cells were fixed in 3.7% formaldehyde for 10 min and permeabilized in 0.2% Triton X-100 for 5 min. After blocking in 1% serum for 20 min, the cells were then incubated with either the preimmune or anti-ELL antisera at a 1:1000 dilution for 1 hr at 37°. After washing in PBS, the cells were incubated with a goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) antibody (Vector Laboratories) for 45 min at 37° and then fixed in 3.75% formaldehyde, quenched in 50 mM NH4, and mounted in Vectashield (Vector Laboratories). The slides were visualized with a Zeiss Axiophot microscope.
RESULTS
Cloning of the Murine Homologue of ELL.
Five cDNA clones were obtained from the screening of the lambda gt11 library. One of the clones had undergone rearrangement and was not pursued further. The two largest clones were 3 kb and were found to be identical. The other two clones were 1.8 kb and 1.2 kb and were completely contained within the larger 3-kb clone. Sequence analysis revealed that one end of the 3-kb clone was highly homologous to ELL beginning at nucleotide 87 of the human sequence. The homology extended throughout the remainder of the open reading frame of ELL and ended at the termination codon. An additional 85 nt upstream of the 3-kb fragment were obtained by the 5′ RACE technique. These sequences contained a Kozak’s consensus start codon after 5 nt of 5′ untranslated sequence. The human sequence contained 12 nt of 5′ untranslated sequence. All clones isolated from the 5′ RACE PCR initiated at the same nucleotide. The combined nucleotide sequence obtained from the cDNA clones and the 5′ RACE has been submitted to the GenBank database.
Sequence Analysis.
The murine Ell sequence contains a predicted open reading frame of 602 aa, slightly smaller than the 621-aa human gene. In addition, 1209 nt of 3′ untranslated sequence were identified in murine Ell. As was found in human ELL, no polyadenylylation signal could be identified in the murine Ell sequence. The murine and human proteins are highly homologous; 85% of the amino acid residues are identical (Fig. 1). Of the 91 amino acid differences, 65 are conservative, 17 are neutral, and only 9 are discrepant. To examine the degree of evolutionary conservation, we compared the murine and human nucleotide sequences with the new diverge program. This showed that of 596 processed codon pairs, 325 pairs have no differences, 215 have one difference, 44 have two differences, and 12 have three differences. The gaps in the predicted proteins and the amino acid differences appear to cluster together at several regions, especially within the central portion of the predicted Ell protein. A 15-aa domain from amino acids 450–465 of Ell shared homology to a region of rat elongin (Fig. 2). No other homology to elongin outside this region was found. In addition, no homology could be identified to other known transcription elongation factors or to other genes deposited in the databases that were searched.
Figure 1.
Comparison of the predicted protein sequence of the murine (upper row) and human (lower row) homologues of ELL. Identical amino acids are indicated by a line, conservative changes by two dots, and neutral changes by one dot.
Figure 2.
Homology of the lysine-rich, hydrophilic region of Ell to a similar region of elongin. Identical amino acids are indicated by a line, conservative changes by two dots, and neutral changes by one dot.
Northern Blot Analysis.
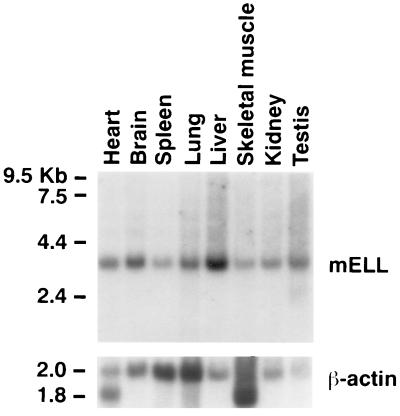
To determine the expression of Ell mRNA in different organs, a murine multiple-tissue Northern blot was hybridized with a 1.2-kb cDNA probe containing the 3′ UTR of Ell (Fig. 3). When the blot was rehybridized with a probe containing the first 700 bp of coding sequences of Ell, an identical pattern of hybridization was observed (data not shown). A single 3.4-kb transcript could be visualized in all tissues examined. No additional bands were observed on prolonged exposure of the autoradiographs. To correct for the amount of RNA loaded in each lane, we measured the intensity of each of the bands on the blot after the hybridization with the actin probe. The highest level of expression was detected in the testis and in the liver. The ratios of the levels of RNA expression relative to that of skeletal muscle and spleen were as follows: testis, 11.5; liver, 6.5; brain, 3.8; kidney, 3.5; heart, 3.2; lung, 1.9; skeletal muscle, 1; and spleen, 1.
Figure 3.
(Upper) Northern blot analysis of multiple murine tissues probed with a 1.2-kb fragment of the 3′ UTR of Ell. (Lower) Hybridization with the actin control. The size of the molecular weight standards are shown on the left.
In Situ Hybridization.
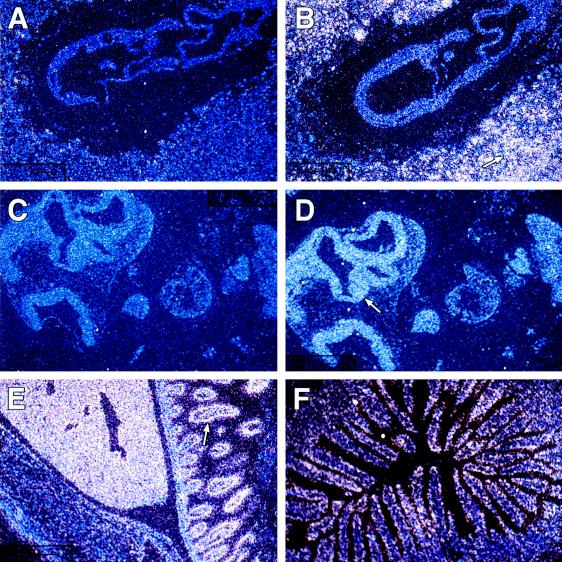
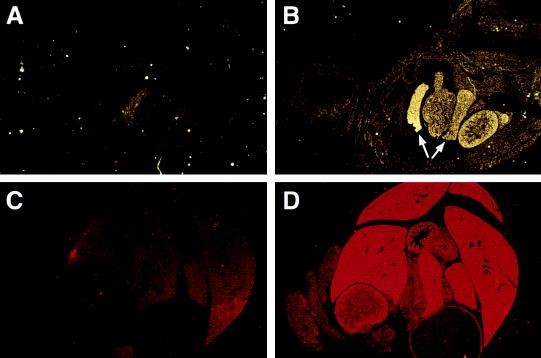
To examine the spatial and temporal pattern of expression of Ell in murine development, in situ hybridization studies were performed on mouse embryos and on adult tissues. On embryonic day 7.5 (E7.5), the antisense probe hybridized diffusely throughout the embryo (Fig. 4 A and B). In addition, high levels of Ell expression were detected in the maternally derived decidual tissue. The sense probe exhibited only minimal background staining on all sections. On E9.5, expression remained diffuse throughout the embryo with slightly more prominent expression in the neuroepithelium (Fig. 4 C and D). At E14.5, diffuse expression throughout the embryo could be detected with the antisense probe compared with the sense control (data not shown). However, by day 16.5, a more specific pattern of expression became evident, with high levels of expression in the liver and gastrointestinal tract (Fig. 5 A and B). The sections from 1-day-old and adult tissues hybridized with the antisense probe also exhibited high levels of expression in the liver and gut (Figs. 4E and 5 C and D). Higher-power magnification revealed that the expression in the liver was in the hepatocytes. In the gut, the hybridization was greatest at the epithelial lining of the intestinal villi (Fig. 4F). This pattern of high expression in the liver and gut with lower level diffuse expression persisting in the remainder of the tissues was first observed at E16.5 and continued until birth, as well as into the adult stage.
Figure 4.
The temporal and spatial pattern of Ell expression during murine development. In situ hybridization studies were performed with radiolabeled Ell 3′ UTR sense (A and C) and antisense (B, D, E, and F) riboprobes. (A and B) Hybridization to longitudinal sections of E7.5 embryonic tissues (×10). The arrow indicates the maternally derived decidual tissue that exhibited high levels of Ell expression. (C and D) Hybridization to longitudinal sections of E9.5 embryonic tissues (×10). Although expression is diffuse throughout the embryo at E9.5, it is slightly more prominent at the neuroepithelium as indicated by the arrow. (E) Hybridization to a longitudinal section of the abdominal cavity of a 1-day-old mouse (×1.25). The abdominal wall is at the lower left and exhibits only minimal Ell expression. In the middle of the panel, a high level of expression is seen in the liver. The arrow indicates expression in the intestine. (F) Hybridization to a cross-section of adult intestine (×10) and illustrates the localization of Ell expression to the intestinal villi.
Figure 5.
In situ hybridization studies were performed with radiolabeled Ell 3′ UTR sense (A and C) and antisense (B and D) riboprobes. (A and B) Hybridization to longitudinal sections of E16.5 embryonic tissues (×1.25). The arrows indicate expression in lobes of the liver. The gastrointestinal tissues, which are adjacent to the liver, also display prominent expression. (C and D) Hybridization to cross-sections of the abdominal cavity of a 1-day-old mouse (×1.25). Prominent expression can be appreciated in the hepatic and gastrointestinal tissues with the antisense probe compared with the sense control.
Immunofluorescence.
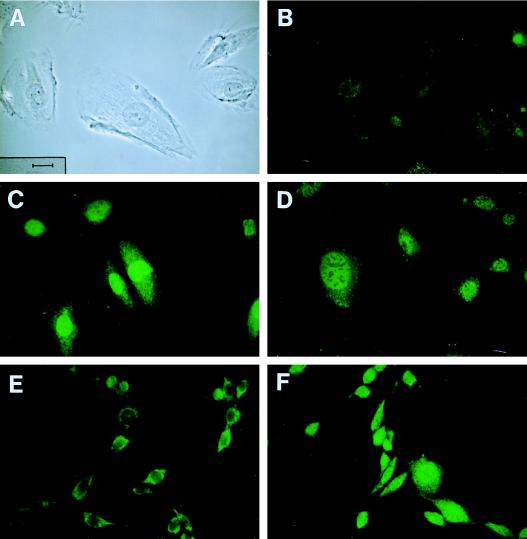
To explore the subcellular localization of the ELL protein, indirect immunofluorescence was performed with both the preimmune and anti-ELL antisera on the A7r5, NIH 3T3, COS-7, and HeLa cell lines (Fig. 6). In these cell lines, the preimmune serum showed a low-level background cytoplasmic staining. With the anti-ELL antiserum, a nuclear signal was detected in all of the cell lines. Although the polyclonal antiserum was generated against the human ELL protein, it also crossreacted with cell lines of primate, murine, and rat origin. The nuclear staining pattern was diffuse but excluded nucleoli. Because of their large size, this was best visualized in the A7r5 cells but could also be seen in the other three cell lines examined. In addition, a perinuclear halo could be visualized in all cell lines but was best appreciated in the A7r5 cells and in subpopulations of larger cells in the NIH 3T3, COS-7, and HeLa cell lines.
Figure 6.
The subcellular localization of ELL. Immunofluorescence studies were performed with preimmune serum (B and E) or a polyclonal antiserum generated to ELL panels (C, D, and F). (A) A phase contrast image of A7r5 cells, which illustrates the size and morphology of these cells. The scale is shown on the lower left and is the same in all panels. (B) Immunofluorescence with the preimmune serum in A7r5 cells, which illustrates the background cytoplasmic staining. (C and D) Immunofluorescence with the ELL antiserum, which illustrates the nuclear localization with exclusion of the nucleoli. (E and F) Immunofluorescence studies with NIH 3T3 cells. The preimmune antiserum again showed only background cytoplasmic staining, whereas the ELL antiserum showed localization to the nucleus and exclusion of the nucleoli. ×40. (Bar length = 50 microns.)
DISCUSSION
When we first identified ELL as a translocation partner gene of MLL in the t(11;19)(q23;p13.1), we could not determine its function based on the amino acid sequence. To characterize the functions of ELL further, we have cloned the murine homologue of ELL. Although murine Ell is slightly smaller than its human homologue, there is a high degree of conservation between amino acid residues. At the nucleotide level, 55% of the potential codon pairs have no differences. Of the 215 codon pairs with a one nucleotide difference, the majority are synonymous changes that do not result in amino acid changes. Thus, this degree of evolutionary conservation suggests that sequence constraints exist within the functional motifs of ELL. We compared the murine and human ELL nucleotide and amino acid sequences to databases using multiple algorithms. As was the case with our searches at the time ELL was cloned, no clear homologies to other genes deposited in the databases could be identified.
Recently, Shilatifard et al. (13) purified a protein from rat liver nuclear extracts that had the capacity to function as an RNA polymerase II elongation factor. Peptide microsequencing revealed that this protein was the rat homologue of ELL. Although only short peptide sequences from the rat are available for comparison to the murine and human sequences, a high level of conservation is apparent. Whereas the sequences previously noted to be conserved between the rat and human were also conserved in the mouse, the sequences that diverged between rat and human sequence also tended to be discrepant between rat and mouse, again suggesting sequence constraints in functional motifs. We did identify a small region within the basic, lysine-rich motif of ELL that shared homology to a region of elongin. The functional significance of this region is unknown and would be valuable to characterize further. No homologies to elongin outside this region or to other known transcription elongation factors were found.
Previously, we found that in multiple-tissue Northern blots hybridized with human ELL probes, a predominant 4.4-kb transcript was expressed along with a 2.8-kb transcript that could be detected in all tissues with prolonged exposure of autoradiographs. In the murine multiple-tissue Northern blots, only a single 3.4-kb band was identified. The source of the additional band on the human Northern blots remains unclear. No evidence for either alternative splicing or of other related genes that might crosshybridize with ELL probes has been obtained. To explore this possibility, the murine Northern blots were probed with both coding sequence and 3′ UTR probes. However, an identical pattern of expression was observed with both probes. A total of 3023 nt of Ell were obtained by screening the cDNA library and from the 5′ RACE. The open reading frame of Ell is 19 aa smaller than its human homologue. Thus, the size difference between mouse and human ELL detected by Northern blot analysis appears to be primarily at the level of the 3′ UTR. We used 5′ RACE to obtain 85 nt of the 5′ end of murine Ell that contained a short 5′ UTR and an initiating methionine. The size of the murine 5′ UTR and the position of the initiating methionine are consistent with our previous 5′ RACE results on the human sequence. After correcting for differences in RNA loading, the level of mRNA expression was found to be highest in the testis and liver. The high level of expression in the liver on the Northern blot is consistent with our data from the in situ hybridization experiments.
To gain insight into the pattern of expression of Ell, we performed in situ hybridization studies in murine embryonic sections and adult tissues. By E7.5, Ell is expressed diffusely. Of all the sections examined, we observed the highest level of expression of Ell, compared with the sense control, in maternally derived decidual tissue. At E14.5, expression within the embryo remained diffuse. However, by E16.5, a specific pattern of high-level expression in the liver and gut became evident that persisted in the adult. This discrete pattern of expression of a general transcription factor was unexpected and suggests that specific genes may be the target of the transcriptional elongation activity of ELL. The expression of Mll in murine embryonic development has previously been studied (17). Mll is expressed widely from E7 and continues throughout development and in adults. Pronounced expression was noted in regions of Hox gene expression such as the embryonic nervous system, in somites, and in somite derivatives. Thus, the normal expression patterns of Mll and Ell are quite divergent.
To determine the subcellular localization of ELL, we generated a polyclonal antiserum to the C terminus of ELL, which was used for immunofluorescence. We obtained similar results in all four cell lines examined. Because these lines were derived from four different species, the crosshybridization of the antibody confirms the high degree of evolutionary conservation of ELL that we identified at the sequence level. We found that ELL is expressed diffusely in the nucleus but excludes nucleoli. A similar nucleoplasmic pattern of expression has previously been noted for RNA polymerase II (16). Because nucleoli are the site of ribosomal RNA synthesis, this suggests that ELL does not function as an elongation factor for RNA polymerase I. The subcellular localization of only one other MLL partner gene, ENL, has been studied. Transient transfections of COS-7 cells with a hemagglutinin-tagged ENL expression construct, followed by immunohistochemical staining with anti-hemagglutinin antibodies, demonstrated only nuclear staining (18). No data have been published on the localization of other transcriptional elongation factors. However, the localization of the VHL protein, which interacts with the elongin gene product, has been characterized (19). VHL exhibits three different patterns of expression: cytoplasmic, cytoplasmic and nuclear, and predominantly nuclear. In the cells with predominantly nuclear localization, the pattern of expression is diffuse with exclusion of nucleoli. The role of elongation factors in either RNA polymerase I or III transcription is not known.
Recent evidence indicates that the VHL tumor suppressor gene binds to the B and C subunits of elongin, thus inhibiting its transcriptional elongation activity. Mutations within VHL may disrupt this regulatory network, leading to aberrant transcription of downstream targets. Thus, the regulation of general elongation factors appears to play a critical role in normal gene expression as well as in tumorigenesis. Because ELL is the first MLL partner gene with a known function, we can now propose models for the potential mechanisms of 11q23 leukemias. At this time, it is not clear if the consequences of (11;19) translocations are due to loss or gain of function for either MLL or for ELL. The fusion of MLL to ELL could potentially inhibit the ability of ELL to serve as an elongation factor for certain critical target genes. Alternatively, the fusion of ELL to MLL could bring the elongation activity of ELL to transcription units regulated by MLL, thus leading to inappropriate expression of these genes. The identification of the normal target genes and interacting proteins for both MLL and ELL will be essential to address these questions.
Acknowledgments
We thank Janet D. Rowley for advice and helpful discussions. This research was supported by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences (M.J.T.), National Institutes of Health Grants HL52094-02 (M.C.S.) and CA42257 (J.D.R.), and the Howard Hughes Medical Institute.
Footnotes
Abbreviations: RACE, rapid amplification of cDNA ends; UTR, untranslated region; GST, glutathione S-transferase; E, embryonic day.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U80227U80227).
Thirman, M. J., Mbangkollo, D., Kobayashi, H., McCabe, N. R., Gill, H. J., Rowley, J. D. & Diaz, M. O. 84th Proceedings of the American Association for Cancer Research, May 19–22, 1993, Orlando, FL.
References
- 1.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thirman M J, Gill H J, Burnett R C, Mbangkollo D, McCabe N R, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg A A, Chaganti R S K, Larson R A, Le Beau M M, Diaz M O, Rowley J D. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 3.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 4.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale R P, Nowell P C, Kuriyama K, Miyazaki Y, Croce C M, Canaani E. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- 7.Bernard O A, Mauchauffe M, Mecucci C, van den Berghe H, Berger R. Oncogene. 1994;9:1039–1045. [PubMed] [Google Scholar]
- 8.Corral J, Forster A, Thompson S, Lampert F, Kaneko Y, Slater R, Kroes W G, van der Schoot C E, Ludwig W-D, Karpas A, Pocock C, Cotter F, Rabbitts T H. Proc Natl Acad Sci USA. 1993;90:8538–8542. doi: 10.1073/pnas.90.18.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tse W, Zhu W, Chen H S, Cohen A. Blood. 1995;85:650–656. [PubMed] [Google Scholar]
- 10.Chaplin T, Ayton P, Bernard O A, Saha V, Della-Valle V, Hillion J, Gregorini A, Lillington D, Berger R, Young B D. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 11.Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H, Huebner K, Berger R, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1994;91:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N J, King G, Rabbitts T H. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 13.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 14.Aso T, Lane W S, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 15.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 16.Chambon P, Gissinger F, Kedinger C, Mandel J L, Meilhac M. In: The Cell Nucleus. Busch H, editor. New York: Academic; 1974. pp. 270–307. [Google Scholar]
- 17.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 18.Rubnitz J E, Morrissey J, Savage P A, Cleary M L. Blood. 1994;84:1747–1752. [PubMed] [Google Scholar]
- 19.Duan D R, Humphrey J S, Chen D Y, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1995;92:459–463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]