Abstract
A two-step gene replacement procedure was developed that generates infectious adenoviral genomes through homologous recombination in Escherichia coli. As a prerequisite, a human adenovirus serotype 5 (Ad5)-derived genome was first introduced as a PacI restriction fragment into an incP-derived replicon which, in contrast to ColE1-derivatives (e.g., pBR322 or pUC plasmids), is functional in a polA mutant of E. coli. Any modification can be introduced at will following two consecutive homologous recombinations between the incP/Ad5 replicon and the ColE1 plasmid. The overall procedure requires only the in vitro engineering of the ColE1-derivative by flanking the desired modification with small stretches of identical sequences. In the first step, a cointegrate between the tetracycline-resistant incP/Ad5 replicon and the kanamycin-resistant ColE1-derivative is selected by growing the polA host in the presence of both antibiotics. Resolution of this cointegrate is further selected in sucrose growth conditions due to the loss of a conditional suicide marker (the sacB gene of Bacillus subtilis) present in the ColE1 plasmid, leading to unmodified and modified incP/Ad5 replicons that can be differentiated upon restriction analysis. Consecutive rounds of this two-step cloning procedure allowed the introduction of multiple independent modifications within the virus genome, with no requirement for an intermediate virus. The potential of this procedure is demonstrated by the recovery of several E1E3E4-deleted adenoviruses following transfection of the corresponding E. coli-derived genomes in IGRP2 cells.
Adenoviruses are small but complex widespread DNA viruses that induce a cytopathic productive cycle in a large variety of somatic cells (for a review, see ref. 1). Forty-nine serotypes have been identified so far in humans. Among them, serotypes 2 (Ad2) and 5 (Ad5) have been used extensively as a basis for the construction of defective gene transfer vehicles for mammalian cells. There are many reasons for this. First, recombinant adenoviruses (RAd) can be prepared to high titers (i.e., 1011 infectious particles/ml), a prerequisite for efficient in vivo delivery. Also, RAd are nonenveloped viruses that are not subjected to complement-mediated inactivation. In addition, Ad2- or Ad5-derived recombinants do not integrate within the host genome, lowering the risk of insertional mutagenesis in the infected cells. Preclinical data accumulated over the past decade indeed demonstrate the potential of RAd for in vivo gene delivery in many types of dividing or quiescent somatic cells from many organs (liver, lung, heart, brain, blood vessels, skeletal muscle cells, etc). Since 1993, RAd have been evaluated in a growing number of phase I clinical trials that target cancers or cystic fibrosis (2). Preclinical and clinical data have underscored the dose-dependent host responses against the virus and virus-infected cells that typically follow administration, clearly calling for improved vectors and technologies (this report) for long-term RAd-mediated gene delivery (C.T., unpublished results).
The Ad2 or Ad5 genetic information is compacted within a linear, 36-kb-long, double-stranded DNA molecule. Both strands of the chromosome are coding, and nearly all transcription units are heavily spliced. The viral transcription units are conventionally referred to as early (E1A, E1B, E2, E3, and E4) and late, depending on their time of expression relative to the onset of viral DNA replication. The pIX and IVa2 transcripts do not belong to either class and are usually referred to as delayed early. The complexity of the virus genome organization renders its manipulation quite difficult and often restricted to certain regions. Indeed, most RAd lack a 3-kb fragment from the E1 region often substituted for the recombinant expression cassette. E1-deleted RAds do not propagate in most human cells and must be amplified in an E1-transcomplementing cell line such as 293 (3) or 911 (4). Accordingly, their genomes retain most (i.e., >80%) of the virus information within a ca. 30-kb DNA molecule.
The in vitro handling of such large DNA molecules is always difficult due to the lack of suitable restriction sites. The construction of E1-deleted RAd is usually accomplished within 293 cells by homologous recombination between a recombinant plasmid corresponding to the left end of the recombinant (i.e., with the transgene replacing the E1 genes), and overlapping DNA of virus origin that has been rendered noninfectious by in vitro restriction. In practice, the recombinant is often diluted with contaminating genomes, especially if it can be counterselected during amplification, and therefore must be purified by consecutive plaquing on 293 cell monolayers. This tedious and time-consuming procedure is not satisfactory to recover clinical grade recombinants because it is not a true cloning process. In addition, it also favors the emergence of replication-competent adenoviruses during virus amplification (5).
We have previously reported the characterization of a 293-derived cell line (IGRP2) that transcomplements the E4 viral function because it expresses the distal moiety of the E4 locus from a dexamethasone-inducible promoter (6). This new packaging cell line was shown to efficiently propagate several E1E4 doubly defective recombinants expressing the Escherichia coli lacZ gene from the Rous sarcoma virus long terminal repeat (RSV LTR). Unfortunately, its plaquing efficiency is below that of 293 with regard to E1-deleted adenoviruses, emphasizing the need for an alternative strategy to recover clonal RAd preparations.
Recently, a complete Ad2 genome has been included as a SnaBI restriction fragment within a yeast artificial chromosome (Ad2 YAC), and infectious recombinant genomes could be constructed from the Ad2 YAC by a two-step gene replacement procedure in Saccharomyces cerevisiae (7). This methodology clearly contributes significant improvements to conventional RAd construction technology. For example, yeast clones harboring the recombinant genomes can be identified by Southern hybridization and truly cloned. Furthermore, multiple modifications can, in principle, be introduced along the virus backbone by recombinational cloning in yeast, with no requirement for intermediate viruses. However, homologous recombination in S. cerevisiae relies on the utilization of DNA linearized at a conveniently located restriction site to target the recombination event within identical sequences (7). It also requires extraction of the YACs from yeast spheroplasts prepared from rather large (e.g., 500 ml) cultures.
To overcome these technological drawbacks, we have developed a clonal genetic technology to construct infectious E. coli-derived RAd genomes (EDRAG). We have taken advantage of the fact that (i) in contrast to ColE1-derivatives—e.g., pBR322 (8) and pUC-derivatives (9)—incP-plasmids accommodate large inserts (10, 11) and are able to replicate in polA mutants of E. coli (12), and (ii) the SacB gene of Bacillus subtilis is a lethal marker for E. coli in the presence of sucrose (13). These properties have been used to construct incP-carried RAd genomes by a two-step recombinational cloning in E. coli. During the first step, a selectable cointegrate is formed by homologous recombination between a ColE1-recombinant plasmid and a resident incP replicon present in E. coli polA, provided that there is homology between the two plasmids. Resolution of this cointegrate by homologous recombination is subsequently selected due to the concomitant loss of the SacB conditional suicide marker. This methodology was used to generate PacI-excisable RAd genomes that are infectious as exemplified by the clonal recovery of E1- and E1E4-defective viruses amplified into 293 and IGRP2 cells, respectively.
MATERIALS AND METHODS
Construction of Recombinant Plasmids.
Recombinant DNA techniques used in this study have been described (14). Plasmids were transformed into TG1 (15), DH5αF′ (CLONTECH), MC1060 (16), or E. coli polA mutants SF800 or C2110 (12). pXL1635 has been described (17), and it derives from pRK290, which harbors an incP replicon (18). When appropriate, the media were supplemented with kanamycin (Km), ampicillin (Ap), spectinomycin (Sp), tetracycline (Tet), or 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) at 50, 100, 100, 12, and 40 mg/liter, respectively.
pCLAI is a pIC19H-derivative (19) that contains the left end of Ad5 (position 1–454, nucleotide numbering is that of Ad5; ref. 20). This plasmid was used for the amplification of the left end of the Ad5 genome (up to position 385) with oligonucleotides 1 (5′-CGGCGGGAATTCTTAATTAACATCATCAATAATATACCTTATTTTGG- 3′) and 2 (5′-CACCACCTGCAGGGTACCACTAGTGTCTCCACGTAAA CGGTCAAAGTC-3′; the adenoviral sequences are underlined). pPY23 is a pIC19H-derivative (19) containing a 0.5-kb BclI–AvrII fragment from plasmid pFG144 (21). This plasmid was used for the amplification of the right end of the Ad5 genome (position 35,517–35,935) with oligonucleotides 1 and 3 (5′-CACCACCTGCAGGGCAGCCTAACAGTCAGCC TTACC-3′; the viral sequences are underlined). The PCR products were digested with EcoRI and PstI, cloned into pUC19 (9), and sequenced. They were cloned together into EcoRI-linearized pUC19 to generate pXL2626, which carries both termini of Ad5 bordered by EcoRI and PacI sites (see italics on oligonucleotide 1 sequence). A 4.5-kb PstI cassette containing the 1.8-kb spectinomycin resistance cassette from pHP45Ω (22) and the sacB gene of B. subtilis, which confers lethality for E. coli grown in the presence of sucrose (13), was cloned into PstI-linearized pXL2626 to generate pXL2636. This plasmid contains a 5.1-kb EcoRI fragment that confers Spr and Sucs, and that harbors both ends of the Ad5 genome within a PacI subfragment. Insertion of this EcoRI fragment into EcoRI-linearized pXL1635 generated pXL2672.
pGY63 is a derivative of pXL2675 [a Kmr derivative of pBKS+ (Stratagene); unpublished data]. It harbors the left inverted terminal repeat (ITR) and the packaging signal (Ψ) (from position 1 to the HinfI site at position 382), part of the pIX gene (from the Sau3A site at position 3446 to the PvuII site at 3968) and, in between, the pCMV-nlslacZ expression cassette. This cassette is composed of the enhancer–promoter from the immediate-early gene of human cytomegalovirus [pCMV; derived from plasmid pCDNA3 (Invitrogen)] and a fragment from pCH-NLS (23) containing the E. coli lacZ gene with a nuclear localization signal (nls). pYJ6 contains the right end of the Ad5 genome (from the TaqI site at position 33,055 but with a deletion extending from the SspI site at position 33,423 to the SmaI site at 35,356) cloned into plasmid pXL2756, a sacB-containing derivative of pXL2675.
pLC1, a derivative of pXL2756, contains the Ad5 sequence from position 1 to 382 and the pIX-encoding sequence (from the BglII site at position 3328 to the NsiI site at position 4419) with an expression cassette, pRSVnls-lacZ, from plasmid pAd.RSVβgal (24) inserted in between. This cassette contains the RSV LTR and expresses an nls–lacZ genetic fusion. pACK2 is another derivative of pXL2756. It harbors the Ad5 sequence from position 1 to 382, followed by a pCMV-driven herpes simplex virus-1 thymidine kinase (HSV1-TK) expression cassette, and Ad5 sequences from position 3447 to 4419.
Selection Protocols for Homologous Recombination.
One hundred nanograms of the pBR322-derived plasmid containing the desired sequences (e.g., pFG144 Apr, pYJ6 Kmr, pGY63 Kmr, pLC1 Kmr, or pACK2 Kmr) was introduced into electrocompetent E. coli SF800 cells, harboring either a Tetr incP-based replicon only containing the adenoviral terminal sequences (pXL2672), or Tetr incP replicons containing recombinant adenoviral genomes (pXL2689, pXL2789, or pXL2811, see below). The first crossing-over between the pBR322- and the incP-derived plasmid generates a cointegrate that is selected on LB agar supplemented with the required antibiotics (Tet+Ap or Tet+Km), and X-Gal, when appropriate. X-Gal was used because the pCMV- and RSV LTR-driven lacZ expression cassettes were functional enough to generate a LacZ+ phenotype in E. coli. A second crossing-over was either selected due to the loss of the sacB gene (Sucr), or by curing the pBR322-derived plasmid. Loss of the sacB gene was selected by plating the cointegrates onto LB plates containing 5% sucrose (13), Tet, and X-Gal when appropriate. Recombinants were reisolated twice on this medium and subsequently maintained on LB Tet plates. When sucrose selection was not possible (e.g., pFG144, as this ColE1 plasmid does not harbor the sacB gene), the cointegrate was introduced into E. coli MC1060 and the bacteria were grown for 80 generations in liquid medium under nonselective pressure. Kms clones were found, and their plasmid content was screened by restriction analysis on plasmid minipreparations.
Cells and Viruses.
Cell culture medium was from GIBCO. IGRP2 (1) and 293 (4) cell lines have previously been described. The RAd generated following transfection of EDRAG is named by the four digits of the transfected plasmid (e.g., transfection of pXL2689 in 293 generates virus Ad2689). E1-defective RAds were recovered in 293 cells by transfecting 10 μg of PacI-digested EDRAG by the Lipofectamine-based procedure (Life Technologies, Gaithersburg, MD). After 10 days, cells were harvested, frozen, thawed three times in a dry ice-ethanol bath, and centrifuged at 3000 × g for 10 min. The cell lysate was added to fresh 293 cells, and a cytopathic effect (CPE) was typically observed after 5 days. E1E4 doubly defective adenoviruses were recovered similarly, except that transfection was carried out in IGRP2 cells as described (6). At the fourth amplification step on fresh IGRP2 cells, a generalized CPE was observed and viral DNA was extracted (25).
Probes for Southern analyses were 32P-labeled by random priming (Amersham) of PCR fragments generated with the following pairs of oligonucleotides: 5′-GGAAGTGACAATTTTCGCGCGG-3′ and 5′-GTCTCCACGTAAACGGTCAAAGTC-3′ to amplify a fragment containing Ψ, and 5′-TCGTTTCTCAGCAGCTGTTG-3′ and 5′-AATACACAGGACCCTCAACGACC-3′ to amplify a fragment containing part of the pIX gene. The E4-specific probe was obtained by random priming of the 336-bp SmaI–SspI fragment from coordinates 33,093–33,423 of the Ad5 genome.
Expression of HSV1-TK was assessed by immunochemistry using a specific rabbit polyclonal antibody (pAbTK41; Rhône-Poulenc Rorer, Vitry-Sur-Seine, France). Briefly, W162 cells (26) were incubated in chamber slides and transduced with Ad3017. After 24 hr, the cells were fixed with 3.5% formaldehyde, washed thoroughly with PBS, and permeabilized with 0.1% Triton X-100. After saturating with 0.5% fetal calf serum, pAbTK41 and fluorescein isothiocyanate-conjugated anti-rabbit IgG (Sigma) were added.
RESULTS
Construction of Ad5 Genomes with E1 and E3 Deletions Bordered by Unique PacI Restriction Sites.
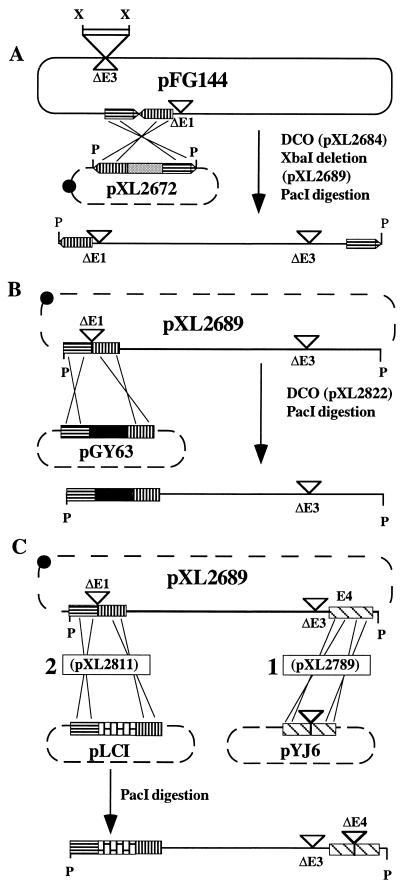
Plasmid pXL2689, from which the RAd genome can be excised by PacI digestion, was constructed as follows (Fig. 1A). pXL2672, corresponding to pXL1635 with a 5.1-kb fragment containing both ends of Ad5 bordered with PacI sites (see Materials and Methods), was transformed into SF800, an E. coli polA mutant. pFG144 (21)—containing an Ad5 genome with the viral termini head to tail, an E1 deletion, and a pBR322-derivative as a replicon inserted in place of an E3 deletion—was then transformed into SF800 harboring pXL2672. Selection for Ap and Tet gave clones in which pFG144 had integrated into pXL2672 by a single crossing-over (Fig. 1A). Counterselection for Sucr and analysis for Sps clones generated pXL2684, which resulted from a second crossing-over. An XbaI deletion removed the pBR322-derived replicon contained within the RAd genome, leading to pXL2689 (Fig. 1A). This plasmid contains an Ad5-based genome carrying deletions within E1 (a SacII deletion extending from positions 354 to 3824) and E3 (a XbaI deletion extending from positions 28,592 to 30,470) (21) and, unlike pFG144, the RAd genome can be excised by PacI-restriction.
Figure 1.
Construction of incP-derived plasmids harboring recombinant Ad5 genomes. (A) Construction of pXL2689. pXL2672 is a pRK290-based incP replicon that contains the right and left Ad5 termini. The only clone obtained during pXL2672 construction contained the expected 5.1-kb EcoRI fragment with the Ad5 termini, together with an additional 4.1-kb EcoRI fragment encompassing the glnE gene of E. coli (GenBank accession no. Z21844Z21844). This plasmid was used further because the presence of this additional fragment did not alter the PacI-mediated excision of the fragment bordered by the viral ITRs. The two consecutive homologous recombinations that led to the construction of plasmid pXL2684 by gene replacement are referred to as DCO (double crossing-over) between pFG144 and pXL2672. XbaI deletion then generated plasmid pXL2689. PacI restriction of pXL2689 produced a RAd genome that contains deletions within the E1 and E3 regions. Vertical and horizontal striped arrows represent the left and right termini, respectively, and the shaded box refers to the sacB gene and the gene conferring Spr. The unshaded boxed area represents pMX2, a 2.2-kb ColE1 (pBR322) derivative carrying an ampicillin-resistance cassette. (B) Construction of pXL2822 by gene replacement (DCO) between pXL2689 and pGY63. Recombinational cloning followed by PacI restriction generates an infectious RAd genome that contains a functional CMV-nlslacZ expression cassette (black box). Vertical and horizontal striped boxes represent the left ITR and Ψ, and sequences encompassing the pIX gene, respectively. (C) Construction of pXL2789 and pXL2811. (1) Gene replacement (DCO) between the Tetr pXL2689 (incP) and Kmr pYJ6 (ColE1) plasmids generated pXL2789, an incP replicon containing an E1−E3−E4− RAd genome. The striped boxes represent E4 sequences. (2) Gene replacement between pXL2789 and pLC1 subsequently generated pXL2811, an incP-based replicon that contains a PacI-excisable E1−E3−E4− recombinant genome containing an RSV LTR/nlslacZ expression cassette (bricked box). Recombinational gene replacement between this incP replicon and pACK2 (ColE1) generates pXL3017, which is identical to pXL2811 except that a pCMV-driven HSV1-TK expression cassette replaces that of pXL2811. Solid lines represent adenoviral sequences and dashed lines refer to non-viral sequences. Crossed lines represent recombination events. The black dot represents the RK2 origin of replication (incP). (▿, deletion; X, XbaI; P, PacI.)
The two-step recombinational procedure was again applied to plasmids pXL2689 (incP) and pGY63 (ColE1), generating pXL2822. This incP replicon is identical to pXL2689 except that it contains a pCMV-nlslacZ expression cassette within a smaller E1 deletion (Fig. 1B).
E. coli-Derived E1E3-Deleted Recombinant Genomes Are Infectious in 293 Cells.
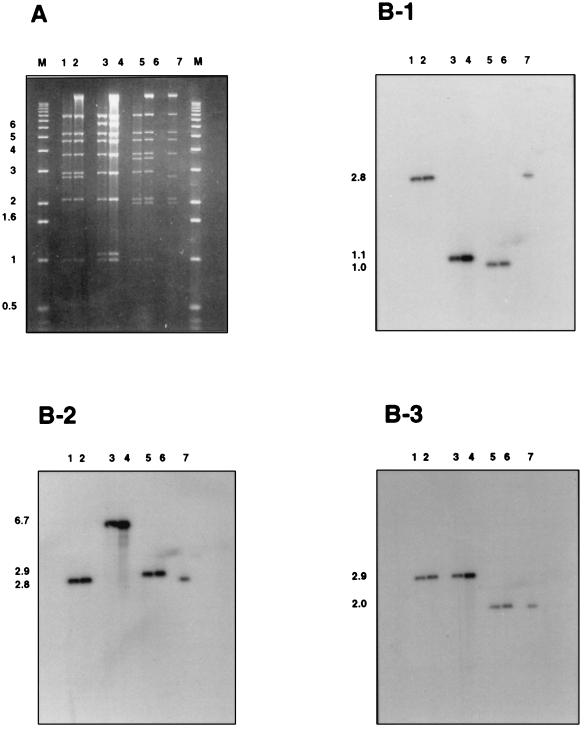
The 30.6-kb and 35.6-kb recombinant genomes generated after PacI restriction of pXL2689 and pXL2822, respectively, exhibit three or five additional nucleotides as compared with normal adenoviral genomes. These E. coli-derived genomes also lack the 55-kDa terminal protein that is covalently linked to their 5′ end and is known to stimulate viral recovery following transfection into permissive cells. PacI-digested pXL2689 and pXL2822 were therefore transfected into 293 cells to assess their infectivity. Although no CPE was evident after transfection, a CPE did appear at the first amplification step. The HindIII restriction patterns of the viruses’ DNA were analyzed and were those expected for viruses Ad2689 and Ad2822, respectively (Fig. 2A). To investigate whether any rearrangement, occurring at a low frequency, was present in the RAd preparations, Southern blot analysis was performed on HindIII-restricted viral DNA with probes from three different regions of the Ad5 genome. In all cases the only hybridizing fragment derived from the RAd genome comigrated with the corresponding hybridizing fragment from the E. coli plasmid (Fig. 2B). As expected, fragments of 2.8 kb and 1.1 kb from Ad2689 and Ad2822, respectively, were the only fragments to hybridize with the Ψ probe. Similarly, the expected 6.7-kb and 2.8-kb fragments from Ad2689 and Ad2822, respectively, were found to hybridize to the probe containing part of the pIX gene. Finally, the 2.9-kb HindIII fragment carrying part of the E4 region was the only fragment to hybridize to the probe derived from the E4 region. Taken together, these results demonstrate that the E1-deleted RAd genomes contained within the PacI restriction fragments were infectious in 293 cells, and that the expected RAds were homogeneous.
Figure 2.
Southern blot analyses of virus DNA as compared with that of their corresponding E. coli-derived plasmid ancestors. (A) Agarose gel electrophoresis (0.8%) of PacI + HindIII-restricted incP replicons and HindIII-restricted viral DNA from cell-transfected extracts. Lanes: M, 1 kb marker; 1 and 2, Ad2689 and pXL2689, respectively; 3 and 4, Ad2822 and pXL2822, respectively; 5 and 6, Ad2811 and pXL2811, respectively; and 7, pXL2789. (B) Autoradiograms from the blot obtained from the gel shown in A hybridized with probes that encompass the packaging sequence Ψ (B-1), the pIX gene (B-2), or the E4 region of Ad5 (B-3). Sizes of the expected fragments are indicated on the left.
Construction of E1E3E4-Deleted Recombinant Adenoviral Genomes in E. coli.
Plasmids containing a recombinant genome deleted in the E1, E3, and E4 regions (extending from the 5′ end of ORF1 to the 3′ end of ORF6 of the E4 region) were constructed by homologous recombination between incP-based pXL2689, pXL2789, and pXL2811 replicons, and pYJ6, pLC1, and pACK2 ColE1-derivatives, respectively, generating pXL2789, pXL2811, and pXL3017 incP-based modified replicons (Fig. 1C). The E. coli clones containing pXL2811 (insertion of the pRSV-nlslacZ cassette in place of E1) turned blue on X-Gal indicator plates, whereas plasmid-less clones or clones with pXL2789 exhibited only a faint coloration, demonstrating a higher expression of lacZ from the pRSV-nlslacZ cassette as compared with that from the endogenous copy of the bacterial chromosome. This property allows ready identification of bacterial clones containing incP-derived replicons in which the pRSV-nlslacZ cassette of pXL2811 has been substituted by a non-lacZ expression cassette by recombinational cloning. For example, clones containing pXL3017, an incP replicon identical to pXL2811 except that a pCMV-driven HSV1-TK expression cassette replaces the pRSV-nlslacZ cassette, exhibited a faint blue phenotype following the two-step recombinational procedure between pACK2 and pXL2811.
E. coli-Derived E1E3E4-Deleted Adenoviral Genomes Are Infectious in IGRP2 Cells.
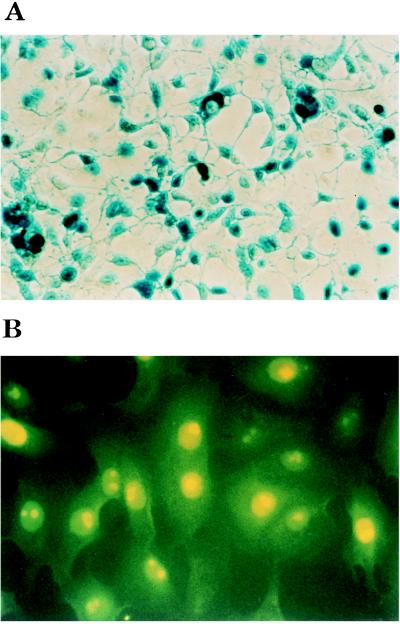
PacI-restricted pXL2811 DNA was transfected into IGRP2 cells (6). A typical, and total, adenovirus-induced CPE was apparent only at the fourth amplification step, indicating a less efficient process than in the case of E1-deleted RAd recovered from 293. The presence of large quantities of a functional Ad2811 virus was further suggested by the observation of numerous β-galactosidase-expressing cells following infection and X-Gal staining (Fig. 3A). Titration of the virus onto IGRP2 cell monolayers confirmed the recovery of normal titers of virus as blue plaques were detected up to the 10−7 dilution after X-Gal staining. HindIII-restricted viral DNA also gave a restriction pattern identical to that obtained from a double PacI–HindIII restriction digest of plasmid pXL2811 (Fig. 2A). Moreover, Southern blot analyses of Ad2811 and pXL2811 identified the same 1-kb, 2.9-kb, and 2-kb fragments when probes derived respectively from Ψ, the pIX gene, and the E4 regions (Fig. 2). No additional fragments were found to hybridize to these probes, indicating that the analyzed viral stock was indeed homogeneous (Fig. 2). The lack of detectable genome heterogeneity in the viral stock clearly indicates that one can take advantage of the clonality of the RAd genomes produced in E. coli to avoid plaque purification of the RAds recovered after transfection of the corresponding genomes into the packaging cells.
Figure 3.
Expression of E. coli β-galactosidase and HSV1-TK in IGRP2 and W162 cells, respectively. (A) IGRP2 cells were infected with Ad2811 and stained with X-Gal at 48 hr postinfection, as described (6). (B) Immunofluorescence analysis of Ad3017-infected W162 with a rabbit serum raised against purified HSV1-TK.
The EDRAG technology has been used to generate many other E1E3E4-deleted RAd genomes (P.B., A.L.R., C.O., and C.T., unpublished results), including pXL3017, which contains a pCMV/HSV1-TK expression cassette. PacI-restricted pXL3017 was transfected into IGRP2 cells and amplified. The RAd genome was extracted by the Hirt procedure and analyzed at the fourth amplification step. Restriction analyses again generated the expected pattern for Ad3017, with no contaminating fragments. This RAd is also functional because in contrast to mock-infected cells, W162-infected cells exhibited a specific dose-dependent nuclear labeling in the presence of the pAbTK41 serum (Fig. 3B).
DISCUSSION
We have developed a recombinational methodology to engineer infectious EDRAG. The EDRAG are plasmid-borne and are modified by two successive rounds of homologous recombination in E. coli. This technology differs from the previously published methods using E. coli because the RAd genomes are entirely constructed in E. coli (i.e., virus emergence does not rely on recombination within the transcomplementing cell line; refs. 27 and 28), and also because the viral genomes do not contain a bacterial origin of replication (21). The EDRAG technology is also extremely powerful. Indeed, plasmid minipreparation analyses and large-scale purification of E. coli-derived plasmids are more easily accomplished than for YAC extracted from S. cerevisiae. For instance, in our standard protocol, 10 μg of PacI-restricted purified plasmid were used for transfection in 293 or IGRP2 cell lines; similar quantities of restricted YAC DNA with the same degree of purity would have been difficult to obtain. The cloned adenoviral genome can be modified using standard E. coli genetic manipulations with restriction digestion and/or homologous recombination. For example, a unique restriction site (such as SwaI or NspI) could be inserted by recombinational cloning within the E1 deletion to facilitate the construction of RAd genomes by direct cloning of compatible recombinant fragments. Moreover, the EDRAG technology has proved rapid and efficient as compared with conventional procedures that require time-consuming plaque purification steps. Because the EDRAG method relies on the transfection of a linear fragment obtained from a bacterial plasmid preparation, the transfected material is clonal and generates a homogenous virus following amplification. Indeed, the different viral preparations were always found to be homogenous by restriction and Southern blot analyses.
We have previously reported the characterization of 293-derived cell lines expressing either the entire E4 locus (IGRP4 cells) or only its distal (ORF6 + ORF7) moiety (IGRP2 cells) from a dexamethasone-inducible promoter (6). Functional analysis has demonstrated that these cell lines allowed the construction of several doubly defective (E3+) deletants expressing the lacZ gene from the RSV LTR (6). However, their construction was a tedious, time-consuming, nonclonal process that required multiple rounds of virus purification on IGRP2-derived cell monolayers (6). The clonal recovery of E1E4 doubly defective RAd expressing transgenes different from lacZ will be even more complex because most viral plaques could not be detected when X-Gal staining was omitted (E.V. and J.F. Dedieu, unpublished data). In this regard, the EDRAG technology will simplify the construction of E1E4 doubly defective RAd. This is an important issue because these vectors should exhibit a reduced emergence of replication-competent adenoviruses during viral amplification, as well as a reduced cytotoxic lymphocyte-mediated clearance of the recipient cells (6). For example, we have used the EDRAG technology to construct many E1E3E4-deleted RAds, including virus Ad3017, which expresses a functional HSV1-TK gene as shown by its ability to confer ganciclovir-dependent toxicity for the infected cells (A. Mahfoudi and M. Janicot, personal communication). The EDRAG technology is indeed simple, rapid, and efficient, and it only requires: (i) the in vitro engineering of small Kmr ColE1 recombinant plasmids, (ii) two selectable homologous recombinations between the ColE1-recombinant and the Tetr lacZ-expressing incP replicon (pXL2811), ultimately leading to the desired incP-based RAd backbone that confers a screenable LacZ− phenotype, and (iii) PacI restriction of the incP-based recombinant prior to transfection within the packaging cells.
Acknowledgments
We thank A. Gillardeaux, I. Loquet, and V. Conard for technical assistance and T. Ciora for synthesis of oligonucleotides. J.-B. Le Pecq is acknowledged for his constant support and interest. We thank D. Lecoq for providing the sacB gene within plasmid pSL301, B. Cameron for critical reading of the manuscript, and A. Mahfoudi for helpful discussion. This work was part of the BioAvenir program supported by Rhône-Poulenc, the French Ministry of Research, and the French Ministry of Industry.
Footnotes
Abbreviations: RAd, recombinant adenovirus(es); Ad2, human adenovirus serotype 2; Ad5, human adenovirus serotype 5; CPE, cytopathic effect; EDRAG, E. coli-derived RAd genome; HSV1-TK, herpes simplex virus-1 thymidine kinase; ITR, inverted terminal repeat; nls, nuclear localization sequence; YAC, yeast artificial chromosome; Ψ, packaging signal; pCMV, human cytomegalovirus; X-Gal, β-d-galactoside; RSV LTR, Rous sarcoma virus long terminal repeat.
References
- 1.Shenk T. In: Virology. Fields B, editor. New York: Raven; 1996. pp. 2111–2148. [Google Scholar]
- 2.Ali M, Lemoine N R, Ring C J A. Hum Gene Ther. 1994;1:367–384. [PubMed] [Google Scholar]
- 3.Graham F L, Smiley J, Russel W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 4.Fallaux F J, Kranenburg O, Cramer S J, Houweling A, van Ormondt H, Hoeben R C, van der Eb A J. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 5.Lochmüller H, Jani A, Huard J, Prescott S, Simoneau M, Massie B, Karpati G, Acsadi G. Hum Gene Ther. 1994;5:1485–1491. doi: 10.1089/hum.1994.5.12-1485. [DOI] [PubMed] [Google Scholar]
- 6.Yeh P, Dedieu J-F, Orsini C, Vigne E, Denèfle P, Perricaudet M. J Virol. 1996;70:559–565. doi: 10.1128/jvi.70.1.559-565.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Proc Natl Acad Sci USA. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 9.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C, Smith C. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 11.Ditta G, Stanfield S, Corbin D, Helinski D R. Plasmid. 1980;13:149–154. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stachel S, An G, Flores C, Nester E. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloomfield I C, Vaughn V, Rest R F, Eisenstein B I. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 14.Maniatis T, Fritsch E, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Gibson, T. J. (1984) Ph.D. thesis (Univ. of Cambridge, Cambridge, U.K.).
- 16.Casadaban M J, Martinez-Arias A, Shapira S K, Chou J. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 17.Crouzet J, Levy-Schil S, Cauchois L, Cameron B. Gene. 1992;110:105–108. doi: 10.1016/0378-1119(92)90451-t. [DOI] [PubMed] [Google Scholar]
- 18.Ditta G, Stanfield S, Corbin D, Helinski D R. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh J L, Erfle M, Wykes E J. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 20.Chroboczek J, Bieder F, Jacrot B. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh-Choudhurry G, Haj-Ahmad Y, Brinkley P, Rudy J, Graham F. Gene. 1986;50:161–171. doi: 10.1016/0378-1119(86)90321-5. [DOI] [PubMed] [Google Scholar]
- 22.Prentki P, Krisch H M. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 23.Nouvel P, Pantier J J, Condamine H. Virology. 1994;204:180–189. doi: 10.1006/viro.1994.1522. [DOI] [PubMed] [Google Scholar]
- 24.Stratford-Perricaudet L D, Makeh I, Perricaudet M, Briand P. J Clin Invest. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg D H, Ketner G. Proc Natl Acad Sci USA. 1983;80:5383–5386. doi: 10.1073/pnas.80.17.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Proc Natl Acad Sci USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bett A J, Haddara W, Prevec L, Graham F L. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]