Abstract
We previously found that gene transduction by adeno-associated virus (AAV) vectors in cell culture can be stimulated over 100-fold by treatment of the target cells with agents that affect DNA metabolism, such as irradiation or topoisomerase inhibitors. Here we show that previous γ-irradiation increased the transduction rate in mouse liver by up to 900-fold, and the topoisomerase inhibitor etoposide increased transduction by about 20-fold. Similar rates of hepatic transduction were obtained by direct injection of the liver or by systemic delivery via tail vein injection. Hepatocytes were much more efficiently transduced than other cells after systemic delivery, and up to 3% of all hepatocytes could be transduced after one vector injection. The presence of wild-type AAV, which contaminates many AAV vector preparations, was required to observe a full response to γ-irradiation. Injection of mice with AAV vectors encoding human clotting factor IX after γ-irradiation resulted in synthesis of low levels of human clotting factor IX for the 5-month period of observation. These studies show the potential of targeted gene transduction of the liver by AAV vectors for treatment of various hematological or metabolic diseases.
The liver is an important target for gene therapy, because of its large size and protein synthetic capacity, and because of the need to target gene transfer to the liver for treatment of diseases involving defects in members of sequential enzymatic pathways that are unique to the liver, such as the urea cycle enzymes. Models for hepatic gene therapy have been developed using retroviral, adenoviral, and DNA-mediated gene transfer, but all of these methods have important limitations. Retroviral vectors can promote long-term expression of genes introduced into the liver after hepatocyte transduction in vitro followed by reimplantation (1) or after direct infection in vivo (2–4). Disadvantages of in vitro gene transfer include the difficulties of cultivation of large numbers of hepatocytes and potential morbidity to the hepatocyte donor from portal vein thrombosis. Direct in vivo gene transfer is limited by the need for liver ablation by partial (70%) hepatectomy (3–5) or by chemical damage (2) to stimulate liver regeneration and cell division necessary for transduction by the retroviral vectors. Adenoviral vectors allow very efficient in vivo gene transfer to the liver in the absence of liver regeneration and can promote very high protein expression levels, but in general, expression is transient due to immune response against adenoviral proteins that are expressed by current adenoviral vectors (6). Receptor-targeted delivery of DNA to the liver can result in prolonged protein production at therapeutically relevant levels, but transfer rates are highly variable and protein expression is often low and transient (7).
Adeno-associated virus (AAV) vectors have a unique set of properties that make them attractive for use in gene therapy (8). Unlike other viral vectors, AAV vectors are derived from a nonpathogenic parvovirus. AAV vectors can integrate in the host genome and allow long-term expression in transduced cells and their progeny. Only the terminal repeats of AAV are required for encapsidation and integration of the viral genome (9, 10), thus all viral genes can be removed from AAV vectors, and vector stocks can be generated by providing AAV and adenovirus helper functions in trans (10). AAV vectors can transduce nondividing cells; however, the rate of transduction is much lower than that for dividing cells in culture (11). Still, long-term expression of tyrosine hydroxylase and β-galactosidase has been reported after AAV vector transduction of nondividing neurons in the rat brain (12), and low-level transduction of neurons in the rat hippocampus was observed after injection of an AAV vector that expressed human placental alkaline phosphatase (AP) (13).
Treatment of cultured cells with agents that affect DNA metabolism, including γ-irradiation and topoisomerase inhibitors, can dramatically improve AAV vector transduction of both dividing and nondividing cells in these cultures (14, 15). For example, the transduction rate achieved in stationary human fibroblasts with a vector encoding AP was 80-fold higher after γ-irradiation (14). In addition, previous γ-irradiation can increase AAV vector transduction of brain epithelium and myotubes in rats (13). The mechanism for increased AAV vector transduction in response to these agents is unknown, but may involve the expression of DNA repair enzymes that allow second-strand synthesis and transcription of the AAV vector genome. Similar increases in transduction rate have been observed after transfer of specific adenovirus genes into cells in culture or in animals, and the stimulation correlates with production of double-stranded forms of the AAV vector (16, 17).
Here we have explored the potential of AAV vectors for hepatic gene transfer. The rate of hepatocyte transduction by an AAV vector expressing a histochemical marker protein was very low in untreated mice, but was enhanced by up to 900-fold after treatment of the mice with γ-irradiation localized to the liver. Tail vein injection of AAV vectors that encode human clotting factor IX (hFIX) into mice resulted in the production of low levels of hFIX in the circulation for the 5-month period of observation.
MATERIALS AND METHODS
Cell Culture.
Human foreskin fibroblasts (18), rat dermal fibroblasts (18), IB3 transformed human airway epithelial cells (19), 293 transformed human kidney cells (20), HeLa human cervical carcinoma cells (21), HepG2 human hepatoma cells (22), and NIH 3T3 thymidine kinase-negative mouse embryo fibroblasts (23) were grown in DMEM with 10% heat-inactivated (30 min at 56°C) fetal bovine serum and 0.9 μg/ml amphotericin B at 37°C in a 10% CO2/air atmosphere.
Vector Construction and Production.
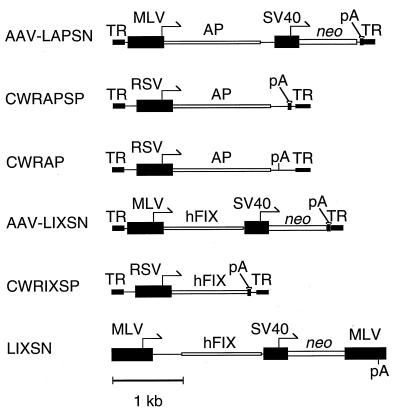
AAV vectors are depicted in Fig. 1. AAV-LAPSN has been described (11). AAV-LIXSN was derived from AAV-LAPSN and contains the hFIX cDNA in place of the AP cDNA. CWRAPSP was constructed from CWRSP (gift of S. Chatterjee, City of Hope, Duarte, CA) by insertion of the AP cDNA between the Rous sarcoma virus promoter and the simian virus 40 polyadenylylation signal. pCWRSP was derived from pCWR-PA (24) by insertion of a simian virus 40 polyadenylylation signal downstream of the polylinker cloning site. CWRIXSP is analogous to CWRAPSP but contains the hFIX cDNA in place of AP. The plasmid pCWRAP (ref. 25; gift of S. Chatterjee) is identical to pCWRAPSP except for the inclusion of slightly more AAV sequence internal to the 3′-terminal repeat, which includes an AAV polyadenylylation signal and no simian virus 40 polyadenylylation signal. The retroviral vector LIXSN has been described (26).
Figure 1.
AAV and retroviral vectors. Arrows indicate promoters, open boxes indicate coding regions, and filled boxes indicate noncoding regions. TR, AAV terminal repeat; MLV, Moloney murine leukemia virus long terminal repeat; AP, alkaline phosphatase; neo, neomycin phosphotransferase; pA, polyadenylylation signal; RSV, Rous sarcoma virus long terminal repeat; hFIX, human FIX. The sequences between the AP coding region and the simian virus 40 promoter in AAV-LAPSN and CWRAPSP are 3′ untranslated sequences from the AP cDNA.
AAV vector stocks were prepared as described (13, 18). The fractions containing the AAV vector were recovered from the cesium chloride gradient and dialyzed against four changes of Ringer’s solution through a 50-kDa molecular mass cutoff dialysis membrane (Spectraphor, Los Angeles) over 16 h at 4°C. The dialyzed vector sample was then concentrated by centrifugation at 1000 × g in a CENTRICON 100 concentrator at 4°C in a Sorvall SS34 rotor to a volume of 1–5 ml. Residual adenovirus was inactivated by heating the vector stock at 56°C for 1 h, and concentrated vector stocks were stored at −70°C until use. The lack of adenovirus contamination was confirmed by plaque assay and by the lack of cytopathic effects of the vector stocks in cell culture, with a sensitivity limit of 1000 plaque-forming units/ml for most vector stocks, and 100 plaque-forming units/ml for some samples. Vector titer was assigned based on the ability to transduce G418 resistance to HeLa cells (AAV-LIXSN) or AP to IB3 cells (AAV-LAPSN, CWRAPSP, and CWRAP). The number of vector DNA-containing particles per AP+ focus-forming unit (ffu) was approximately 300:1 for CWRAPSP and 500:1 for CWRAP by Southern blot analysis of extracted virion DNA, and the number of vector DNA-containing particles per colony-forming unit (cfu) was approximately 10,000:1 for AAV-LIXSN and 1000:1 for AAV-LAPSN. For wild-type AAV, the number of viral DNA-containing particles per infectious unit was 100:1.
Vector stocks were tested for wild-type AAV content by using a replication center assay. Cells of the 293 cell line were trypsinized, and 30,000 cells were plated per well in 96-well plates. Twenty-four hours later various dilutions of vector stock were added to each well along with adenovirus 5 (multiplicity of infection approximately 80 by adenovirus plaque assay on 293 cells). Thirty hours later 100 μl of PBS containing 12.5 mM ethylenediaminetetraacetic acid was added to each well to suspend the cells, which were aspirated onto nylon membrane filters. Cells were lysed, and cellular DNA was bound to nylon membrane filters as described for bacterial colonies (27). In brief, circular filters were placed on 3MM blotting paper (Whatman) saturated with 0.5 M sodium hydroxide and 1.5 M sodium chloride for 5 min, and then neutralized on 3MM blotting paper saturated with 1 M Tris·HCl (pH 7.0)/2× standard saline citrate for 5 min twice, and crosslinked to the filter. Filters were hybridized to a radiolabeled AAV genomic probe, and foci of replicated AAV DNA from individual cells were detected by autoradiography.
Transduction of Primary Hepatocytes in Mice.
All animal experiments were carried out in accordance with institutional and National Institutes of Health guidelines. Viruses were injected directly into the lobes of the liver or through the tail vein of C57BL/6J mice (The Jackson Laboratory). General anesthesia and laparotomy were necessary to allow direct injection, as described (2). The AAV vector stock was diluted in Ringer’s solution to a final volume of 0.4–0.5 ml. Mice were sacrificed on day five or six by halothane overdose. Tissues were perfused with fixative for 5 min, sectioned 2–3 mm thick, and fixed with 2% paraformaldehyde for 3 h (or 3.7% formaldehyde overnight where specified), washed in four changes of PBS over a 4-h period, heat inactivated at 70°C for 2 h (to inactivate endogenous AP activity), and stained for AP at room temperature overnight in 100 mM Tris (pH 8.5)/100 mM NaCl/50 mM MgCl2/1 mg/ml nitro blue tetrazolium/0.1 mg/ml 5-bromo-4-chloro-3-indolyl phosphate.
Topoisomerase inhibitors were administered intraperitoneally at the time of AAV vector injection. Irradiation of the abdomen was performed 2–16 h before virus injection. The cranium, chest, and lower extremities were shielded by 2-cm-thick lead to protect the lungs and marrow cavities.
Quantitation of Transduced Hepatocytes.
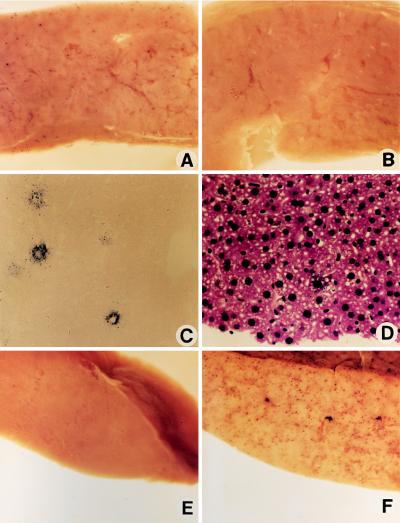
AP+ foci were counted in 0.25-cm2 fields in a standard grid at ×20 magnification. Ten fields were averaged for each liver, and the mean and SD for each group were calculated. We analyzed thin sections of liver to calculate the proportion of transduced hepatocytes. The number of AP+ hepatocytes in a nuclear fast red-stained section (Fig. 2C) was divided by the number of hepatocyte nuclei in a hematoxylin- and eosin-stained section (Fig. 2D) to give the proportion of hepatocytes that were transduced. Specifically, the average number of transduced hepatocytes for 10 fields at 63× power was divided by the average number of hepatocytes in 10 fields at 1000× and divided by 252 to account for the difference in magnification.
Figure 2.
AP staining of the liver after the specified treatment. (A) CWRAPSP at 5 × 105 ffu and 18 Gy of γ-irradiation. (×12.) (B) CWRAPSP at 5 × 105 ffu. (×12.) (C) CWRAPSP at 5 × 105 ffu and 18 Gy. (Nuclear fast red; ×74.) (D) CWRAPSP at 5 × 105 ffu and 18 Gy. (Hematoxylin and eosin; ×74.) (E) AAV-LIXSN at 106 cfu and 17 Gy. (×12.) (F) CWRAP at 5 × 106 ffu and 17 Gy. (×12.)
Quantitative PCR.
Vector DNA was quantitated by competitive amplification as described (18). Total cellular DNA (1 μg) from mouse liver was amplified in a 50-μl reaction (0.01% gelatin/50 mM KCl/10 mM Tris·HCl, pH 8.5/2 mM MgCl2, 0.1 mM each dNTP/0.1 mM [α-32P]dTTP/0.1 mM [α-32P]dCTP/2.5 units of Taq DNA polymerase/200 ng of each primer). The primer sequences were AP exon 4, 5′-AACCAGTGCAACACGACA-3′ and AP-exon 5, 5′-TGGTGGTCACCACTCCCAC-3′. The primers were chosen to recognize regions of highest similarity between the human placental AP sequences (nt 874–892 and nt 1023–1041) (28) and the mouse embryonic AP sequences (nt 1185–1202 and nt 1327–1345) (29). Indicated amounts of virion DNA were added to 1 μg of negative-control mouse liver DNA for generation of a standard curve. Samples were subjected to 25 cycles of amplification with each cycle consisting of 1 min at 95°C, 45 s at 55°C, and 45 s at 72°C. Products were resolved by electrophoresis in nondenaturing 10% polyacrylamide gels, detected by autoradiography of the dried gel, and quantitated by phosphorimaging.
hFIX Assays.
Blood samples for hFIX measurement were handled as described (26), and hFIX was quantitated by ELISA as described (30). Antibodies were obtained from Sigma (mouse monoclonal anti-hFIX and horseradish peroxidase-conjugated goat anti-rabbit) and Boehringer Mannheim (rabbit anti-hFIX antiserum). Normal human plasma was obtained from Arthur Thompson (Puget Sound Blood Center, Seattle, WA) and discarded after four freeze–thaws.
RESULTS
AAV Vector Transduction of Hepatocytes in Mice Is Dramatically Increased by Previous Irradiation.
Mice received tail vein injections of 5 × 105 ffu of the AAV vector CWRAPSP (Fig. 1), which contains an AP cDNA linked to a Rous sarcoma virus promoter. Irradiated animals received a sublethal dose of 18 Gy of γ-irradiation localized to the liver before vector injection. Livers were stained for AP activity 5 days after vector injection. Transduction events were distributed uniformly throughout the liver in all animals, and we observed a dramatic increase in hepatocyte transduction in irradiated mice (Fig. 2A) compared to unirradiated mice (Fig. 2B). The transduction rate after γ-irradiation was 900-fold higher than that observed in unirradiated mice (1450 ± 500 vs. 1.6 ± 2.8 AP+ foci per cm2, respectively; three mice in each group). Equivalent results were obtained with a similar dose of another vector, AAV-LAPSN (Fig. 1), in which the AP cDNA is linked to a Moloney murine leukemia virus promoter (1420 ± 360 foci per cm2 with 17 Gy of irradiation vs. 1.6 ± 4.0 AP+ foci per cm2 without irradiation; two mice in each group). An irradiated mouse that received a vector expressing hFIX, AAV-LIXSN (Fig. 1) did not have AP+ foci in the liver (Fig. 2E), nor did irradiated mice that received an AAV vector expressing β-galactosidase or no vector (not shown). Analysis of thin sections of the liver revealed that the transduced cells were primarily hepatocytes (Fig. 2C). The effects of radiation toxicity, such as hepatocyte necrosis and hepatic venule and sinusoid dilatation, were not observed in hematoxylin- and eosin-stained sections (Fig. 2D).
We next compared the hepatocyte transduction rate obtained by direct injection of an AAV vector into the liver with that obtained by tail vein injection of the vector. Mice were exposed to 18 Gy of γ-irradiation and received 106 ffu of AAV-LAPSN. The lobes of the liver that were injected blanched completely, indicating that the vector stock probably entered the vasculature. The number of transduced hepatocytes in each case was similar (36 ± 8 AP+ foci per cm2 by hepatic injection vs. 36 ± 7 AP+ foci per cm2 by tail vein injection, three animals in each group). The transduction rate observed in this experiment was lower than in previous experiments, in part due to the use of formaldehyde fixative in this experiment that significantly reduced the AP staining. Thus, expression of an AAV vector in the liver was as efficient by tail vein injection as by direct injection into the liver. A similar result previously has been observed for adenovirus vector transduction of hepatocytes, where tail vein injection, portal vein injection, or direct injection of the liver promoted similar rates of hepatocyte transduction (31).
We examined AAV vector transduction of hepatocytes after treatment of mice with agents other than irradiation that also stimulate AAV vector transduction in cell culture. Mice were treated with various amounts of γ-irradiation or with different topoisomerase inhibitors before injection of the AP expression vector CWRAPSP. Data were pooled from mice that received the vector by tail vein injection or by direct injection and were equivalent for both methods under each condition. In all cases, AP staining was distributed uniformly throughout the liver. The greatest induction occurred with 17 Gy of irradiation, resulting in a 130-fold increased transduction rate compared with that of unirradiated animals (Table 1). Etoposide (40 mg/kg) increased liver transduction approximately 17-fold, while camptothecin (5 mg/kg) did not significantly increase liver transduction (Table 1). Thus pretreatment of mice with γ-irradiation resulted in the highest levels of AAV vector transduction, but pretreatment with a topoisomerase inhibitor also resulted in a large increase in the transduction rate.
Table 1.
AAV vector transduction after treatment of mice with various agents that affect DNA metabolism
| Treatment | AP+ foci/cm2 | Fold increase |
|---|---|---|
| None | 15 ± 10 | 1 |
| Camptothecin | 34 ± 38 | <3 |
| Etoposide | 260 ± 130 | 17 |
| 8 Gy of γ-irradiation | 310 ± 200 | 21 |
| 12 Gy of γ-irradiation | 540 ± 180 | 36 |
| 17 Gy of γ-irradiation | 1970 ± 450 | 130 |
Mice received 1 × 105 ffu of CWRAPSP and the indicated treatment on day 1, and livers were fixed with 2% paraformaldehyde and stained for AP on day 6. Mean numbers of AP+ foci per cm2 ± SD are reported for three or four mice in each group. The number of mice injected by tail vein (t) or direct liver injection (d) for each treatment was as follows: none (t = 1, d = 3), 17 Gy (t = 3), 12 Gy (t = 3), 8 Gy (t = 2, d = 1), camptothecin (t = 1, d = 2), and etoposide (t = 1, d = 3).
The Delivery of AAV Vectors Is Liver-Targeted and Efficient After Intravenous Injection.
The highest frequency of transduction after treatment with γ-irradiation or topoisomerase inhibitors occurred in liver. Staining of tissues other than liver for AP revealed very few transduced cells (<40 AP+ foci per cm2 in the lung or kidney after injection of up to 5 × 106 ffu of CWRAPSP, data not shown). Increased vector transduction in the lung after γ-irradiation would have been underestimated, because the chest was shielded from exposure to radiation, but systemic treatment with etoposide increased AP staining in the liver without any increase in lung staining. It is likely that the AAV vector was carried to tissues other than the liver by the circulation, because adenovirus vectors are efficiently delivered to the lung, and to a lesser extent to the spleen and kidney, after tail vein injection (31).
In an attempt to maximize AAV vector transduction of hepatocytes in vivo, we administered 5 × 106 ffu of an AP expression vector (CWRAP) to each of two mice by tail vein injection after 17 Gy of localized hepatic irradiation. CWRAP was chosen for this experiment because it yielded the highest titer vector stocks. The transduction rate in liver was high (Fig. 2F), and about 3% of hepatocytes were transduced based on comparison of the number of transduced hepatocytes to the total number of hepatocytes per microscope field (see Materials and Methods). No evidence of acute toxicity to the mice was observed after vector administration nor was there evidence of liver toxicity by microscopic examination of the liver. The proportion of transduced hepatocytes in mice that were exposed to 18 Gy of γ-irradiation and received 5 × 105 ffu of CWRAPSP was 0.2% (Fig. 2 C and D), showing a nearly linear dose response.
An AAV Vector Encoding hFIX Allows Long-Term Expression in Vivo.
An AAV vector encoding hFIX and neomycin phosphotransferase was constructed (AAV-LIXSN, Fig. 1), and hFIX production from cultured cells exposed to the vector and selected for G418 resistance was analyzed (Table 2). As previously reported for retroviral vectors expressing hFIX (26), hFIX production was higher from primary skin fibroblasts from humans and rats than from established cell lines, including HepG2 human hepatoma cells. hFIX production by primary skin fibroblasts was 2- to 3-fold lower after AAV-LIXSN transduction than after transduction with the retroviral vector LIXSN. Vector RNA levels were proportional to hFIX protein production by phosphorimager analysis of Northern blots of total RNA from transduced cells containing the AAV or retroviral vectors (not shown).
Table 2.
hFIX production in various cells types
| Cell type | hFIX production, ng per 106 cells per day
|
|
|---|---|---|
| AAV-LIXSN vector | LIXSN vector | |
| HeLa | 56 ± 15 | |
| NIH 3T3 | 22 ± 6 | |
| HepG2 | 24 ± 15 | |
| HFF | 520 ± 170 | 960 ± 210 |
| RDF | 450 ± 50 | 1460 ± 290 |
Transduced cells were selected for G418 resistance and were grown to 80–90% confluence before changing the medium and collecting a sample for hFIX ELISA 24 h later. Human foreskin fibroblasts (HFF) and rat dermal fibroblasts (RDF) are primary fibroblasts, while the other cell types are continuous cell lines. Each value represents the mean ± SD for two experiments except the value for NIH 3T3 cells, which is the mean of duplicate samples in one experiment.
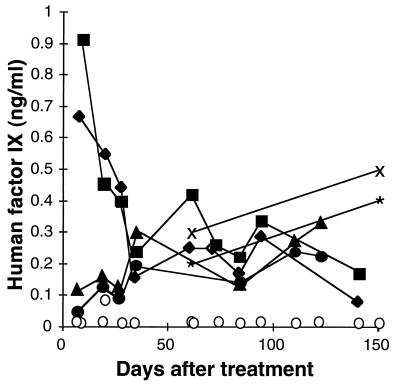
Production of hFIX in mice was analyzed after transduction by AAV vectors in vivo. Mice were exposed to γ-irradiation localized to the liver and were injected with 3 × 105 or 106 cfu of AAV-LIXSN, or 2 × 109 vector-containing particles of another vector expressing hFIX, CWRIXSP. Up to 0.9 ng/ml hFIX was detected in mouse plasma, and hFIX levels of 0.1–0.5 ng/ml persisted for more than 20 weeks (Fig. 3). Mice that were irradiated and received an AAV vector encoding AP served as negative controls in parallel ELISAs (Fig. 3). The limit of detection was 0.02 ng/ml hFIX, and hFIX vector-treated mice had at least 5-fold and generally more than 10-fold greater hFIX levels than negative controls in every assay. Thus, long-term expression of hFIX was possible after transduction of hepatocytes with AAV vectors.
Figure 3.
Plasma levels of hFIX in mice after exposure to γ-irradiation and injection of AAV vectors. Negative-control mice ○ received 17 Gy of irradiation and 5 × 105 ffu of CWRAPSP by tail vein injection. Two mice received 1 × 106 cfu of the AAV-LIXSN vector by tail vein or direct injection and 17 Gy of irradiation (direct injection = ▪ and tail vein injection = ♦), two mice received 3 × 105 cfu of the AAV-LIXSN vector by direct injection and 18 Gy of irradiation (X and ∗), and two mice received approximately 2 × 109 particles of CWRIXSP by tail vein injection and 17 Gy irradiation (▴ and •).
Wild-Type AAV Is Required for Maximal Gene Transfer Rates.
Subsequent testing of the AAV vector stocks used in the foregoing experiments revealed significant contamination by wild-type AAV that was introduced as an inadvertent contaminant of the adenovirus stock used for AAV vector preparation. Use of an AAV-free adenovirus stock for AAV vector preparation reduced the level of AAV contamination to ≤5% of vector levels, but we have been unable to totally eliminate AAV contamination, presumably due to recombination between vector and AAV helper (pAAV/Ad) (10) plasmids. Using a CWRAPSP vector stock with only 0.25% AAV contamination, we found poor transduction of liver after administration of 1 × 105 ffu of the vector with or without 17 Gy of previous γ-irradiation (0.8 ± 1.6 and 4.8 ± 4.0 AP+ foci per cm2, respectively; three mice in each group). In a reconstruction experiment, we added various amounts of AAV to a vector stock that already contained 6% AAV and confirmed that the presence of AAV was required for efficient liver transduction after irradiation (Table 3).
Table 3.
Wild-type AAV is required for maximal liver transduction by an AAV vector after γ-irradiation
| AAV iu/ml
|
AP+ foci/cm2 | Increase, -fold |
|---|---|---|
| AP+ ffu/ml | ||
| 0.06 | 160 ± 80 (n = 3) | 1 |
| 0.11 | 530 ± 200 (n = 3) | 3.3 |
| 50 | 1040 ± 220 (n = 2) | 6.5 |
Mice received vector stocks containing the indicated ratio of AAV infectious units (iu, as determined by an AAV replication center assay) relative to CWRAPSP ffu. Each mouse received 5 × 105 ffu of CWRAPSP supplemented with various amounts of wild-type AAV2 by tail vein injection after 17 Gy of localized irradiation. Livers were fixed and stained for AP on day 6 after treatment. Results are mean numbers ± SD.
Enhanced Transduction Following Irradiation Was Not the Result of Vector DNA Amplification.
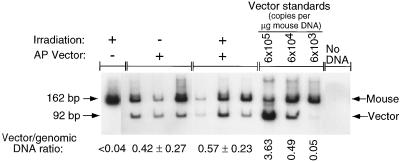
We previously found that increased transduction of stationary human fibroblasts in culture with an AAV vector after γ-irradiation was not due to amplification of vector DNA (14). We performed quantitative PCR of vector DNA from mouse liver DNA to examine this issue after in vivo transduction. Genomic DNA was purified from livers of the mice that had received AAV vectors by tail vein injection with or without 17 Gy of localized irradiation to the liver (Table 1). PCR primers that hybridized to exons 4 and 5 of the mouse and human placental AP genes were used to amplify the human placental AP cDNA in CWRAPSP (92 bp), and equivalent sequences plus intron 4 from mouse genomic DNA (162 bp). The mouse genomic fragment served as an internal standard for the efficiency of amplification in each reaction. The ratio of vector DNA to genomic DNA in livers from mice that received 1 × 105 ffu of CWRAPSP and no irradiation was not significantly different from that of otherwise identically treated animals that received 17 Gy of irradiation (Fig. 4), although the number of transduced hepatocytes increased 130-fold in irradiated animals (Table 1). The vector stock used here contained an excess of wild-type AAV. No AP signal was detected in liver DNA from a negative-control mouse that received 17 Gy of irradiation and 1 × 106 cfu of an AAV vector that did not contain the AP cDNA (Fig. 4). The signal for virion DNA increased as a linear function of the amount of virion DNA added to negative-control mouse liver DNA (Fig. 4), indicating that quantitation was accurate for the range of signals observed for mouse liver DNA samples. Therefore, the increase in transduction after γ-irradiation was due to increased expression from vector DNA, and not to replication or spread of the AAV vector.
Figure 4.
Quantitative PCR of vector DNA in mouse liver DNA. Amplification of vector DNA and mouse genomic DNA was performed to quantify the relative levels of AP vector DNA for the following samples. Mice were treated with 1 × 105 ffu of CWRAPSP, either with or without previous exposure to 17 Gy of γ-irradiation. Liver DNA for a mouse injected with AAV-LIXSN and exposed to 17 Gy served as a negative control for vector DNA amplification. The indicated amounts of virion DNA were added to 1 μg of the negative-control sample to generate standards for vector quantitation. No DNA was added to the last sample. Signals were quantified with a phosphorimager. The signal for the vector DNA (92 bp) was normalized to the signal for mouse genomic DNA (162 bp) to correct for varying efficiencies of amplification. The ratio of vector (cDNA) signal to genomic signal and the SD for each condition are shown.
DISCUSSION
We have demonstrated long-term expression of hFIX from mouse hepatocytes after an intravenous infusion of AAV vector stocks and view this method as a potential strategy for hepatic gene therapy. The targeted delivery of AAV vectors to the mouse liver by a simple tail vein injection after localized γ-irradiation resulted in levels of transduction similar to those achieved by retroviral vectors after partial hepatectomy. Delivery of the vector to the liver by the venous circulation was similar in efficiency to direct injection, as has been observed for adenovirus vectors (31). The level of transduction in the liver was up to 3% of hepatocytes and approached the efficiency of transduction observed in cultured cells. These results indicate that AAV vectors may allow liver gene therapy by a simple intravenous infusion after treatment with an agent to stimulate vector transduction.
The presence of wild-type AAV was necessary to allow increased transduction of hepatocytes by AAV vectors in vivo after γ-irradiation. Fifty percent of the maximum increase in transduction occurred for a vector stock containing only 11% wild-type AAV. On first analysis it would seem that very few hepatocytes would contain both vector and AAV genomes to allow detection of the effect of wild-type AAV on transduction, given that a mouse liver contains 3 × 107 hepatocytes (5) and the vector stock contained only 5 × 105 ffu of the vector and a smaller amount of AAV. However, AAV vector stocks contain many more vector DNA-containing particles than transducing units (300-fold more in this example), and many of these vector genomes enter the cell nucleus (18). Similarly, the ratio of genome-containing particles to infectious units is 100:1 for our wild-type AAV virus stock. Thus, consideration of the number of vector and AAV DNA-containing particles shows that many hepatocytes could contain both vector and AAV DNA.
The likely role of AAV in increasing transduction of irradiated cells is to provide Rep protein, which facilitates second-strand synthesis and vector integration. Demonstration of AAV replication in cells treated with UV irradiation and other mutagenic agents (32) indicates that Rep is synthesized after DNA damage in the absence of adenovirus helper functions. If this is the mechanism of increased AAV vector transduction, providing Rep alone might allow efficient transduction of hepatocytes in vivo without the need for coinfection with AAV. However, it should be noted that AAV itself had no apparent toxic effects on the animals studied here, nor is AAV infection associated with disease in humans despite its high frequency in childhood (33–35). Thus it may be acceptable to use wild-type AAV to stimulate AAV vector transduction for human gene therapy.
Efficient hepatocyte transduction by AAV vectors required local hepatic irradiation, and risks associated with irradiation include malignancy and radiation toxicity to the liver. For patients undergoing bone marrow transplantation after marrow ablation by total body γ-irradiation (9–16 Gy) and chemotherapy, the risk for new malignancy was about 1% and the risk of severe radiation toxicity to the liver was about 9% (36, 37). The risk for hepatic cancer was only 0.03%, indicating that localized liver irradiation would pose a low risk for malignancy. A greater concern is the risk for severe radiation toxicity to the liver; however, the risk was decreased at lower radiation doses (12 versus 16 Gy) and did not occur in patients receiving autologous as opposed to allogeneic marrow, suggesting that severe radiation toxicity would not occur in humans receiving localized hepatic irradiation in a gene therapy setting. Given these risks, specific applications of AAV vectors for gene therapy in the liver could include metabolic disorders, such as branched-chain ketoaciduria or urea cycle disorders, in which partial replacement of a deficient enzyme in the liver might be therapeutic and where current therapy is inadequate (38, 39). To allow more general use of AAV vectors for hepatic gene therapy, other treatments that mimic the effects of irradiation to stimulate AAV vector integration and expression must be developed.
The level of hFIX in plasma of normal humans is about 5 μg/ml, but levels of about 100 ng/ml would prevent chronic disease (40). The plasma level of hFIX in mice transduced with the AAV vector was between 0.1 and 1 ng/ml, similar to levels achieved with a retroviral vector in dogs (3) and well below the therapeutic range. However, several factors indicate that hFIX levels produced by AAV vectors can be increased into the therapeutic range. Substitution of liver-specific promoters for the viral promoters in retroviral vectors resulted in up to 70-fold higher activity in hepatocytes in vivo compared with the murine leukemia virus long terminal repeat promoter (4, 41), and addition of intronic and 3′ untranslated sequences from the hFIX gene also increased hFIX production from an adenoviral vector by more than three orders of magnitude (42). These sequences could be included in AAV vectors to increase protein production. Increasing the amount of AAV vector administered is also likely to result in increased hFIX secretion, although production methods would have to be improved to provide sufficient amounts of vector to treat humans. We found that increasing the dose of an AAV vector encoding AP from 5 × 105 ffu to 5 × 106 ffu resulted in a proportional increase in the number of hepatocytes transduced, from 0.2% to 3%, indicating that we are still in a linear portion of the dose–response curve and that further increases in transduction are likely with higher vector doses.
Alternatively, current levels of protein production that we have achieved would suffice for treatment of other diseases. For example, neutropenia in humans is treated by daily injections of granulocyte colony-stimulating factor (G-CSF), and we have shown that delivery of G-CSF at levels above 100 pg/ml by skeletal muscle cells transduced with a retroviral vector encoding G-CSF causes significant elevation in neutrophil counts in rats (43). We have constructed an AAV vector encoding G-CSF, and preliminary results show serum G-CSF levels of 200 to >1000 pg/ml and markedly increased neutrophil counts in mice receiving this vector after hepatic irradiation. Thus AAV vectors may provide a method for liver-specific gene therapy involving a simple nonsurgical treatment with radiation and intravenous injection of the vector.
Acknowledgments
We are grateful to Arthur Thompson and Yifan Dai for information on hFIX assays, and Kelly McIntyre, Feng Chen, Shall Jue, and Mary Howland for excellent technical assistance. This work was supported by Grants HL41212, DK47754, and HL03100 from the National Institutes of Health. D.D.K. was supported by the Judith Graham Pool Fellowship from the National Hemophilia Society.
Footnotes
Abbreviations: AAV, adeno-associated virus; hFIX, human clotting factor IX; AP, alkaline phosphatase; ffu, focus-forming unit; cfu, colony-forming unit; G-CSF, granulocyte colony-stimulating factor.
References
- 1.Grossman M, Raper S E, Kozarsky K, Stein E A, Engelhardt J F, Muller D, Lupien P J, Wilson J M. Nat Genet. 1994;6:335–341. doi: 10.1038/ng0494-335. [DOI] [PubMed] [Google Scholar]
- 2.Kaleko M, Garcia J V, Miller A D. Hum Gene Ther. 1991;2:27–32. doi: 10.1089/hum.1991.2.1-27. [DOI] [PubMed] [Google Scholar]
- 3.Kay M A, Rothenberg S, Landen C N, Bellinger D A, Leland F, Toman C, Finegold M, Thompson A R, Read M S, Brinkhous D M, Woo S L C. Science. 1993;262:117–119. doi: 10.1126/science.8211118. [DOI] [PubMed] [Google Scholar]
- 4.Rettinger S D, Kennedy S C, Wu X, Saylors R L, Hafenrichter D G, Flye M W, Ponder K P. Proc Natl Acad Sci USA. 1994;91:1460–1464. doi: 10.1073/pnas.91.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kay M A, Li Q, Lui T J, Leland F, Toman C, Finegold M, Woo S L. Hum Gene Ther. 1992;3:641–647. doi: 10.1089/hum.1992.3.6-641. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Furth E E, Nunes F A, Gonczol E, Berencsi K, Wilson J M. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perales J C, Ferkol T, Beegen H, Ratnoff O D, Hanson R W. Proc Natl Acad Sci USA. 1994;91:4086–4090. doi: 10.1073/pnas.91.9.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzyczka N. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 9.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samulski R J, Chang L-S, Shenk T. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell D W, Miller A D, Alexander I E. Proc Natl Acad Sci USA. 1994;91:8915–8919. doi: 10.1073/pnas.91.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O’Malley K L, During M J. Nat Genet. 1994;8:148–153. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 13.Alexander I E, Russell D W, Spence A M, Miller A D. Hum Gene Ther. 1996;7:841–850. doi: 10.1089/hum.1996.7.7-841. [DOI] [PubMed] [Google Scholar]
- 14.Alexander I E, Russell D W, Miller A D. J Virol. 1994;68:8282–8287. doi: 10.1128/jvi.68.12.8282-8287.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell D W, Alexander I E, Miller A D. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher K J, Gao G, Wietzman M D, DeMatteo R, Burda J F, Wilson J M. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrari F K, Samulski T, Shenk T, Samulski R J. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbert C L, Alexander I E, Wolgamot G M, Miller A D. J Virol. 1995;69:1473–1479. doi: 10.1128/jvi.69.3.1473-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeitlin P L, Lu L, Rhim J, Cutting G, Stetten G, Kieffer K A, Craig R, Guggino W B. Am J Respir Cell Mol Biol. 1991;4:313–319. doi: 10.1165/ajrcmb/4.4.313. [DOI] [PubMed] [Google Scholar]
- 20.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 21.Gey G O, Coffman W D, Kubicek M T. Cancer Res. 1952;12:264. [Google Scholar]
- 22.Skelly J, Copeland J A, Howard C R, Zuckerman A J. Nature (London) 1979;282:617–618. doi: 10.1038/282617a0. [DOI] [PubMed] [Google Scholar]
- 23.Wei C M, Gibson M, Spear P G, Scolnick E M. J Virol. 1981;39:935–944. doi: 10.1128/jvi.39.3.935-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podsakoff G, Wong K K, Jr, Chatterjee S. J Virol. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher-Adams G, Wong K K, Jr, Podsakoff G, Forman S J, Chatterjee S. Blood. 1996;88:492–504. [PubMed] [Google Scholar]
- 26.Palmer T D, Thompson A R, Miller A D. Blood. 1989;73:438–445. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 198–199. [Google Scholar]
- 28.Knoll B J, Rothblum K N, Longley M. J Biol Chem. 1988;263:12020–12027. [PubMed] [Google Scholar]
- 29.Manes T, Glade K, Ziomek C A, Millan J L. Genomics. 1990;8:541–554. doi: 10.1016/0888-7543(90)90042-s. [DOI] [PubMed] [Google Scholar]
- 30.Dai Y, Roman M, Naviaux R K, Verma I M. Proc Natl Acad Sci USA. 1992;89:10892–10895. doi: 10.1073/pnas.89.22.10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith T A G, Mehaffey M G, Kayda D B, Saunders J M, Yei S, Trapnell B C, McClelland A, Kaleko M. Nat Genet. 1993;5:397–402. doi: 10.1038/ng1293-397. [DOI] [PubMed] [Google Scholar]
- 32.Yalkinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 33.Blacklow N R, Hoggan M D, Kapikian A Z, Austin J B, Rowe W P. Amer J Epidemiol. 1968;89:368–378. doi: 10.1093/oxfordjournals.aje.a120897. [DOI] [PubMed] [Google Scholar]
- 34.Blacklow N R, Hoggan M D, Sereno M S, Brandt C D, Kim H W, Parrott R H, Chanock R M. Amer J Epidemiol. 1971;94:359–366. doi: 10.1093/oxfordjournals.aje.a121331. [DOI] [PubMed] [Google Scholar]
- 35.Parks W P, Boucher D W, Melnick J L, Taber L H, Yow M D. Infect Immun. 1970;2:716–722. doi: 10.1128/iai.2.6.716-722.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bearman S I, Appelbaum F R, Buckner C D, Peterson F B, Fisher L D, Clift R A, Thomas E D. J Clin Oncol. 1988;6:1562–1568. doi: 10.1200/JCO.1988.6.10.1562. [DOI] [PubMed] [Google Scholar]
- 37.Witherspoon R P, Fisher L D, Schoch G, Martin R, Sullivan K M, Sanders J, Deeg H J, Doney K, Thomas D, Storb R, Thomas E D. N Engl J Med. 1989;321:784–789. doi: 10.1056/NEJM198909213211203. [DOI] [PubMed] [Google Scholar]
- 38.Chuang D T, Shih V E. In: Metabolic and Molecular Bases of Inherited Disease. 7th Ed. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1239–1277. [Google Scholar]
- 39.Brusilow S W, Horwich A L. In: Metabolic and Molecular Bases of Inherited Disease. 7th Ed. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1187–1232. [Google Scholar]
- 40.Thompson A R. Thromb Haemostasis. 1991;66:119–122. [PubMed] [Google Scholar]
- 41.Hafenrichter D G, Wu X, Rettinger S D, Kennedy S C, Flye M W, Ponder K P. Blood. 1994;84:3394–3404. [PubMed] [Google Scholar]
- 42.Kaleko M, Kayda D, Skhuja K, Mehaffey M, McClelland A. J Cell Biochem. 1995;21A:366. [Google Scholar]
- 43.Bonham L, Palmer T, Miller A D. Hum Gene Ther. 1996;7:1423–1429. doi: 10.1089/hum.1996.7.12-1423. [DOI] [PubMed] [Google Scholar]