Abstract
Activation of the c-Abl protein tyrosine kinase by certain DNA-damaging agents contributes to down-regulation of Cdk2 and G1 arrest by a p53-dependent mechanism. The present work investigates the potential role of c-Abl in apoptosis induced by DNA damage. Transient transfection studies with wild-type, but not kinase-inactive, c-Abl demonstrate induction of apoptosis. Cells that stably express inactive c-Abl exhibit resistance to ionizing radiation-induced loss of clonogenic survival and apoptosis. Cells null for c-abl are also impaired in the apoptotic response to ionizing radiation. We further show that cells deficient in p53 undergo apoptosis in response to expression of c-Abl and exhibit decreases in radiation-induced apoptosis when expressing inactive c-Abl. These findings suggest that c-Abl kinase regulates DNA damage-induced apoptosis.
The product of the c-abl gene is a nonreceptor tyrosine kinase that is localized to the nucleus and cytoplasm (1–3). c-Abl shares certain structural feature with the Src family tyrosine kinases and, in addition, contains nuclear localization signals (4), a DNA-binding domain (2), actin-binding domains (5, 6), and proline-rich sequences (7, 8). The SH3 domain of c-Abl binds to Abl-interactor (Abi) proteins 1 and 2 (9, 10), and the protein tyrosine phosphatase SHPTP1 (11). Abi-1, Abi-2, and SHPTP1, as well as the c-Crk adaptor protein (7, 8) and RNA polymerase II (12, 13), have been identified as c-Abl substrates.
The functional role of c-Abl has been unclear. Mice with homozygous disruption of c-abl are born runted and die after several weeks (14, 15). Overexpression of c-Abl is associated with growth arrest in G1 phase by a mechanism that is p53 dependent (16–18). Other studies have shown that suppression of growth by c-Abl is dependent on both p53 and the retinoblastoma (Rb) protein (19). c-Abl associates with Rb (20), and overexpression of kinase-defective c-Abl abrogates Rb-induced growth arrest (21). Recent work has demonstrated that c-Abl is activated by ionizing radiation and certain other DNA-damaging agents (22). Activated c-Abl binds to p53 and regulates ionizing radiation-induced G1 growth arrest by a p53-dependent, p21-independent mechanism (23). The activation of c-Abl by exposure to ionizing radiation also contributes to the induction of Jun kinase (SAPK/JNK) (22) and p38 MAPK (24). Activation of SAPK and p38 MAPK pathways has been associated with induction of apoptosis (25, 26).
The present studies have addressed the involvement of c-Abl in ionizing radiation-induced apoptosis. The results demonstrate that overexpression of c-Abl induces apoptosis and that cells deficient in c-Abl kinase exhibit resistance to apoptosis induced by irradiation.
MATERIALS AND METHODS
Cell Culture.
MCF-7 cells and mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco’s modified Eagle’s medium (Sigma) containing 10% heat-inactivated fetal bovine serum (Sigma), 2 mM l-glutamine, 10 units/ml penicillin, and 100 μg/ml streptomycin. Null pSRαMSVtkneo, pSRαMSVc-Abltkneo, and pSFαMSVc-AblK(290)Rtkneo or N6Neo vectors were introduced into cells by Lipofectamine (GIBCO/BRL) or by infection with virus-free supernatant. Stable lines were established by selection in G418. Cells were treated with ionizing radiation at room temperature with a Gammacell 1000 (Atomic Energy of Canada) with a 137Cs source emitting at a fixed rate dose of 0.76 Gy/min.
Cell Survival Assay.
Cells were exposed to the indicated doses of ionizing radiation and seeded at a density of 100, 1000, or 10,000 per well. Colony formation was assayed at 10 days after treatment with ionizing radiation. For staurosporine (STSP), cells were seeded at 100, 1000, or 10,000 per well 12 h prior to the treatment. Cells were incubated with indicated doses of STSP for 1 h and washed, and then colony formation was assayed at 10 days. Colonies containing more than 50 cells were scored as positive.
Apoptosis Assays.
Terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assays were performed with ApopTag, fluorescein (Oncor catalog no. S7111-KIT). DNA content was assessed by staining ethanol-fixed cells with propidium iodide and monitoring by FACScan (Becton Dickinson). Numbers of cells with sub-G1 DNA content were determined with a modfit lt program (Verity Software House, Topsham, ME).
Immunoblot Analysis.
Cells were lysed in 1% Nonidet P-40 lysis buffer. After boiling in loading dye, cellular proteins were resolved by SDS/PAGE, transferred to nitrocellulose filters, and analyzed with anti-Abl (Ab-3, Oncogene Science), anti-p53 (Ab-1, Oncogene Science), or anti-p21 (sc-397, Santa Cruz Biotechnology). Proteins were detected with an ECL system (Amersham).
RESULTS AND DISCUSSION
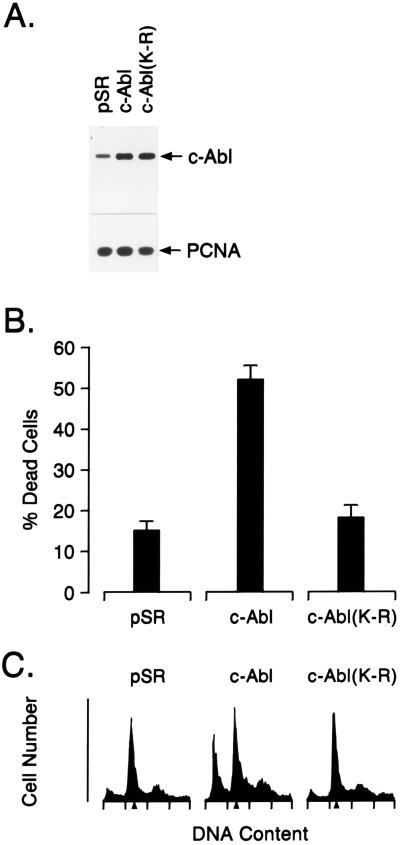
To determine whether c-Abl affects the induction of apoptosis, we transfected MCF-7 cells with vectors expressing wild-type c-Abl or a kinase-inactive K(290)R mutant (16) (Fig. 1A). Expression of wild-type c-Abl, but not c-Abl(K-R), was associated with increased cell death compared with that associated with transfection of the empty vector (Fig. 1B). The c-Abl-transfected cells also exhibited increases in apoptosis as assessed by the appearance of cells with sub-G1 DNA content (Fig. 1C). By contrast, there was no evidence for increased apoptosis in cells transfected with c-Abl(K-R) (Fig. 1C).
Figure 1.
Overexpression of c-Abl induces apoptosis. MCF-7 cells were transfected with 8 μg of control pSRαMSVtkNeo, c-Abl, or c-Abl(K-R) vector. (A) Cell lysates were subjected to immunoblot analysis with anti-Abl and anti-proliferating cell nuclear antigen (PCNA). (B) Cells were assayed for death by trypan blue exclusion at 48 h after transfection. Results are expressed as mean ± SEM of three independent transfections. Transfection efficiency as determined by cotransfection with a pSV-β-gal vector was 40–45%. Cell death for nontransfected MCF-7 cells was approximately 1%. (C) Cells were assayed for DNA content by flow cytometry at 48 h after transfection. Transfection with pSR, c-Abl, and c-Abl(K-R) resulted in 4.5% ± 0.9%, 29.4% ± 4.6%, and 4.1% ± 1.0% (mean ± SEM, three independent experiments) cells with sub-G1 DNA content, respectively.
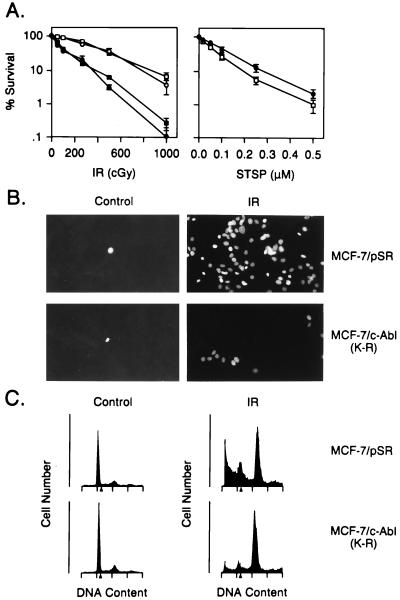
To assess the role of c-Abl in apoptosis induced by DNA damage, we used MCF-7 cells that stably express the dominant-negative c-Abl(K-R) (23). MCF-7/c-Abl(K-R) cells demonstrated resistance to ionizing radiation-induced killing as compared with wild-type and MCF-7/pSR cells (Fig. 2A). Similar results were obtained with two independently isolated MCF-7-c-Abl(K-R) clones (Fig. 2A Left). By contrast, the MCF-7/c-Abl(K-R) cells exhibited little resistance to treatment with staurosporine, which induces killing by DNA damage-independent mechanisms (27, 28) (Fig. 2A Right). TUNEL assays were used to confirm that the radiation-induced cell death was by apoptosis (Fig. 2B). Exposure of MCF-7/pSR cells to ionizing radiation resulted in increased staining, while there was less apoptosis in irradiated MCF-7/c-Abl(K-R) cells (Fig. 2B). In addition, c-Abl(K-R) expression decreased the appearance of irradiated cells with sub-G1 DNA content (Fig. 2C).
Figure 2.
c-Abl(K-R) transfectants are resistant to ionizing radiation-induced apoptosis. (A) Wild-type MCF-7 (▪), MCF-7/pSR (•), and MCF-7/c-Abl(K-R) (clones a and b; □ and ○) cells (23) were exposed to the indicated doses of ionizing radiation (IR) and assayed for colony formation at 10 days (Left). MCF-7/pSR (•) and MCF-7/c-Abl(K-R) (□) cells were exposed to staurosporine (STSP) for 1 h, washed, and then assayed for colony formation at 10 days (Right). Results (mean ± SEM of three experiments) are expressed as the percentage clonogenic survival relative to untreated cells. (B) MCF-7/pSR and MCF-7/c-Abl(K-R) cells were treated with 5 Gy of ionizing radiation (IR). The cells were stained in TUNEL assays at 120 h after irradiation. (×400.) (C) DNA content was analyzed at 120 h after irradiation with 5 Gy.
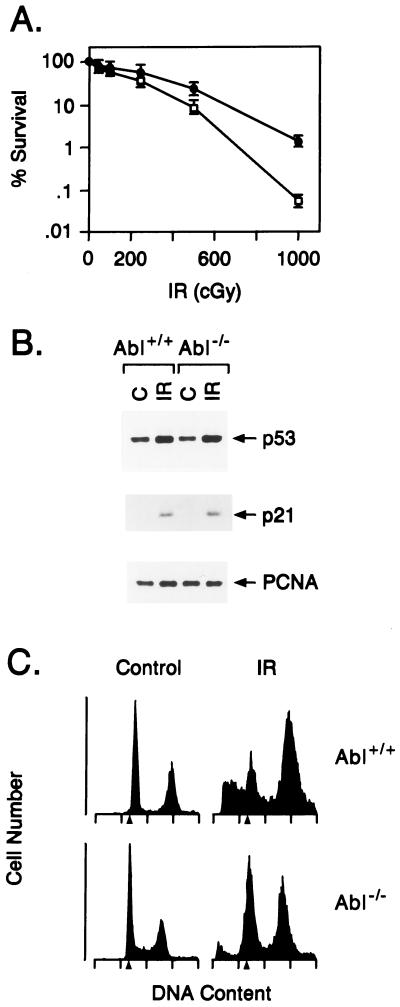
To confirm the link between c-Abl kinase and apoptosis, we studied MEFs deficient in c-Abl (abl−/− mice with targeted disruption of the c-abl gene) (15). The Abl−/− cells demonstrated resistance to the killing effects of ionizing radiation as compared with wild-type MEFs (Fig. 3A). DNA damage increases p53 levels by a posttranscriptional mechanism (29). Both cell types responded to ionizing radiation with induction of p53 (Fig. 3B), indicating that c-Abl is dispensable for increases in p53 expression induced by DNA damage. However, the Abl−/− cells exhibited resistance to ionizing radiation-induced apoptosis. More than 20% of the wild-type MEFs, as compared with about 10% of the Abl−/−, cells exhibited a sub-G1 DNA content after exposure to ionizing radiation (Fig. 3C). These findings indicated that loss of c-Abl kinase abrogates DNA damage-induced apoptosis.
Figure 3.
c-Abl kinase is involved in radiation-induced apoptosis. (A) Wild-type (Abl+/+; ○) and Abl−/− (•) MEFs (15) were exposed to the indicated doses of ionizing radiation (IR) and assayed for colony formation after 10 days. (B) Abl+/+ and Abl−/− MEFs were exposed to 0 or 5 Gy and collected at 3 h. Cell lysates were immunoblotted with anti-p53 and anti-p21 antibodies. (C) Abl+/+ and Abl−/− MEFs were exposed to 0 or 10 Gy and collected at 120 h for analysis of DNA content. Irradiation of the Abl+/+ and Abl−/− cells resulted in 17.3% ± 4.3% and 8.9% ± 2.1% with sub-G1 DNA, respectively.
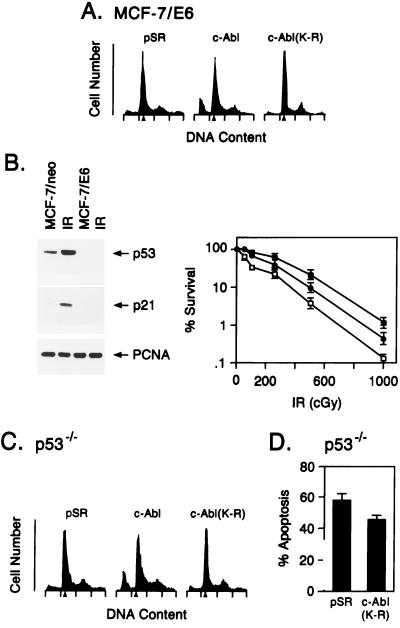
Since c-Abl binds to p53 in vitro and in vivo (23) and loss of p53 function decreases the induction of apoptosis by DNA damage (30, 31), we prepared cells (MCF-7/E6) that stably express the human papillomavirus E6 protein to promote degradation of p53 (32). Transfection of wild-type, but not inactive, c-Abl into MCF-7/E6 cells was associated with induction of apoptosis (Fig. 4A). Treatment of MCF-7/neo cells with ionizing radiation resulted in a 3.2-fold increase in p53 levels and induction of the p53 effector p21, while increases of p53 and p21 levels were attenuated in irradiated MCF-7/E6 cells (Fig. 4B Left). Ionizing radiation-induced killing of MCF-E6 cells was decreased compared with that obtained for MCF-7/pSR cells (Fig. 4B Right). Moreover, stable expression of both E6 and c-Abl(K-R) resulted in further increases in radioresistance (Fig. 4B Right). These results suggest that induction of apoptosis by c-Abl is at least in part p53 independent. To extend the relationship of c-Abl and p53 in apoptosis, we transiently transfected p53−/− fibroblasts with wild-type c-Abl and c-Abl(K-R). Wild-type c-Abl, and not c-Abl(K-R), induced apoptosis in the p53−/− cells (Fig. 4C). The p53−/− fibroblasts were also stably transfected with c-Abl(K-R) to assess radiosensitivity of these cells. The p53−/−/c-Abl(K-R) cells failed to respond to ionizing radiation with activation of c-Abl kinase (data not shown). The p53−/−/c-Abl(K-R) cells also exhibited resistance to ionizing radiation-induced apoptosis as compared with the p53−/−/pSR cells (Fig. 4D).
Figure 4.
c-Abl kinase regulates irradiation-induced apoptosis by p53-dependent and -independent mechanisms. (A) MCF-7/E6 cells were transfected with 8 μg of pSR, c-Abl, or c-Abl(K-R) vector. Cells were harvested at 48 h and assessed for DNA content. Transfection efficiency as determined by cotransfection with pSV-β-gal vector was 40–45%. (B) (Left) MCF-7/pSR (lanes 1 and 2, counting from the left) and MCF-7/E6 (lanes 3 and 4) were irradiated with 5 Gy (lanes 2 and 4). Lysates prepared at 3 h were subjected to immunoblotting with anti-p53 (Ab-6; Oncogene Science) or anti-p21 (Ab-1; Oncogene Science). (Right) MCF-7/pSR (□), MCF-7/E6 (•), and MCF-7/E6/c-Abl(K-R) (▪) cells were irradiated with the indicated doses. Colony formation was assessed at 10 days. (C) p53−/− MEFs (33) were transfected with 8 μg of pSR, c-Abl, or c-Abl(K-R) vector. Transfection efficiency as determined by cotransfection of pSV-β-gal vector was 15–20%. Cells were harvested at 48 h and assessed for DNA content. (D) p53−/− MEFs stably tranfected with pSR vector or c-Abl(K-R) were treated with 10 Gy of ionizing radiation and collected at 120 h for assessment of DNA content. The results (mean ± SEM of three experiments) are expressed as the percentage of apoptotic cells with sub-G1 DNA.
The responsiveness of irradiated cells to undergo G1 arrest (29) or apoptosis (34–36) has been associated with expression of wild-type p53. c-Abl interacts with p53 in irradiated cells and contributes to radiation-induced G1 arrest by a p53-dependent mechanism (23). Mutant forms of p53 have demonstrated that G1 arrest and apoptosis are effected through distinct p53-dependent pathways (37–39). The present studies in MCF-7/E6 and p53−/− cells that express c-Abl(K-R) provide evidence for a c-Abl-dependent, p53-independent, pathway that regulates DNA damage-induced apoptosis. In addition, comparison of radiation-induced killing of MCF-7/c-Abl(K-R) and MCF-7/E6/c-Abl(K-R) cells suggests that c-Abl may also regulate cell death pathways by p53-dependent mechanisms. The basis for involvement of the c-Abl kinase on radiation-induced apoptosis and killing is unclear. In this context, none of the known substrates of c-Abl has been linked to apoptosis. c-Abl has been implicated in DNA recombination and perhaps repair (40). c-Abl also regulates radiation-induced stress signaling pathways (22, 24) that have been implicated in apoptosis (25, 26). While mechanistic studies are now needed, the present findings indicate that c-Abl has a pro-apoptotic function and that loss of c-Abl kinase confers radioresistance.
Acknowledgments
This investigation was supported by Grant CA55241 awarded by the National Cancer Institute.
Footnotes
Abbreviations: MEFs, mouse embryo fibroblasts; TUNEL, terminal deoxynucleotidyltransferase-mediated UTP end labeling.
References
- 1.Wang J Y J. Curr Opin Genet Dev. 1993;3:35–43. doi: 10.1016/s0959-437x(05)80338-7. [DOI] [PubMed] [Google Scholar]
- 2.Kipreos E T, Wang J Y J. Science. 1992;256:382–385. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- 3.McWhirter J R, Wang Y J. Mol Cell Biol. 1991;11:1785–1792. [Google Scholar]
- 4.Van Etten R A, Jackson P, Baltimore D. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 5.McWhirter J R, Wang J Y J. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Etten R A, Jackson P K, Baltimore D, Sanders M C, Matsuddaira P T, Janmey P A. J Cell Biol. 1994;124:325–340. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feller S M, Knudsen B, Hanafusa H. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren R, Ye Z-S, Baltimore D. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Alin K, Goff S P. Genes Dev. 1995;9:2583–2597. doi: 10.1101/gad.9.21.2583. [DOI] [PubMed] [Google Scholar]
- 10.Dai Z, Pendergast A M. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 11.Kharbanda S, Bharti A, Pei D, Wang J, Pandey P, Ren R, Weichselbaum R, Walsh C T, Kufe D. Proc Natl Acad Sci USA. 1996;93:6898–6901. doi: 10.1073/pnas.93.14.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baskaran R, Dahmus M E, Wang J Y J. Proc Natl Acad Sci USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duyster J, Baskaran R, Wang J Y J. Proc Natl Acad Sci USA. 1995;92:1555–1559. doi: 10.1073/pnas.92.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartzberg P L, Stall A M, Hardin J D, Bowdish K S, Humaran T, Boast S, Harbison M L, Robertson E J, Goff S P. Cell. 1991;65:1165–1176. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 15.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 16.Sawyers C L, McLaughlin J, Goga A, Havilik M, Witte O. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 17.Mattioni T, Jackson P K, van Huijsduijnen O B-H, Picard D. Oncogene. 1995;10:1325–1333. [PubMed] [Google Scholar]
- 18.Goga A, Liu X, Hambuch T M, Senechal K, Major E, Berk A J, Witte O N, Sawyers C L. Oncogene. 1995;11:791–799. [PubMed] [Google Scholar]
- 19.Wen S-T, Jackson P K, Van Etten R A. EMBO J. 1996;15:1583–1595. [PMC free article] [PubMed] [Google Scholar]
- 20.Welch P J, Wang J Y J. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 21.Welch P J, Wang J Y J. Mol Cell Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharbanda S, Ren R, Pandey P, Shafman T D, Feller S M, Weichselbaum R R, Kufe D W. Nature (London) 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 23.Yuan Z M, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D. Nature (London) 1996;382:272–275. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 24.Pandey P, Raingeaud J, Kaneki M, Weichselbaum R, Davis R, Kufe D, Kharbanda S. J Biol Chem. 1996;271:23775–23779. doi: 10.1074/jbc.271.39.23775. [DOI] [PubMed] [Google Scholar]
- 25.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 26.Johnson N L, Gardner A M, Diener K M, Lange-Carter C A, Gleavy J, Jarpe M B, Minden A, Karin M, Zon L I, Johnson G L. J Biol Chem. 1996;271:3229–3237. doi: 10.1074/jbc.271.6.3229. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson M D, Burne J F, King M P, Miyashita T, Reed J C, Raff M C. Nature (London) 1993;361:365–369. doi: 10.1038/361365a0. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zelenski N G, Yang J, Sakai J, Brown M S, Goldstein J L. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
- 29.Kuerbitz S, Plunkett B, Walsh W, Kastan M. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J M, Bernstein A. Proc Natl Acad Sci USA. 1993;90:5742–5746. doi: 10.1073/pnas.90.12.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 32.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 33.Lowe S W, Jacks T, Housman D E, Ruley H E. Proc Natl Acad Sci USA. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wylie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 35.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 36.Lowe S W, Ruley H E, Jacks T, Housman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 37.Friedlander P, Haupt Y, Prives C, Oren M. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polyak K, Waldman T, He T-C, Kinzler K W, Vogelstein B. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 39.Rowan S, Ludwig R L, Haupt Y, Bates S, Lu X, Oren M, Vousden K H. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y-Y, Wang L C, Huang M S, Rosenberg N. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]