Abstract
Epstein–Barr virus (EBV) latent membrane protein 1 (LMP1) is essential for transforming primary B lymphocytes into lymphoblastoid cell lines. EBV recombinants with LMP1 genes truncated after the proximal 45 codons of the LMP1 carboxyl terminus are adequate for transformation. The proximal 45 residues include a domain that engages the tumor necrosis factor receptor associated factors (TRAFs). We investigated the importance of the TRAF binding domain by assaying the transforming ability of recombinant EBV genomes with a deletion of LMP1 codons 185–211. This mutation eliminates TRAF association in yeast and in lymphoblasts but does not affect LMP1 stability or localization. Specifically mutated recombinant EBV genomes were generated by transfecting P3HR-1 cells with overlapping EBV cosmids. Infection of primary B lymphocytes resulted in cell lines that were coinfected with an LMP1Δ185–211 EBV recombinant and P3HR-1 EBV, which has a wild-type LMP1 gene but is transformation defective due to another deletion. Despite the equimolar mixture of wild-type and mutated LMP1 genes in virus preparations from five coinfected cell lines, only the wild-type LMP1 gene was found in 412 cell lines obtained after transformation of primary B lymphocytes. No transformed cell line had only the LMP1Δ185–211 gene. An EBV recombinant with a Flag-tagged LMP1 gene passaged in parallel segregated from the coinfecting P3HR-1. These data indicate that the LMP1 TRAF binding domain is critical for primary B lymphocyte growth transformation.
Keywords: NF-κB, TRAF
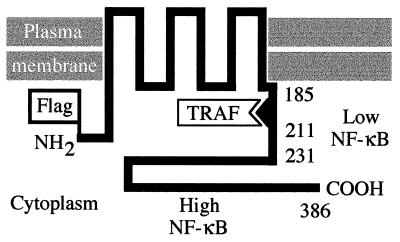
Epstein–Barr virus (EBV) latent membrane protein 1 (LMP1) is expressed in lymphoblastoid cell lines (LCLs) transformed by EBV in vitro, in EBV-associated lymphoproliferative disease, in EBV-associated Hodgkin disease, and in the preneoplastic lesions of nasopharyngeal carcinoma (1, 2). The first 24 and last 200 residues of LMP1 are in the cytoplasm, whereas the intervening 162 residues constitute six membrane-spanning domains separated by peptide turns (see Fig. 1; refs. 3 and 4). Biochemical and recombinant EBV genetic analyses indicate that the membrane-spanning domains of LMP1 enable it to aggregate in the plasma membrane, that primary B lymphocyte transformation into LCLs depends on this aggregation, that the carboxyl terminus (CT) is essential for transformation, and that the amino terminus is not a mediator of transformation (5–7). EBV recombinants that express an LMP1 truncated after the first 45 residues of the CT are able to growth-transform B lymphocytes. However, the transformed cells are not as robust in outgrowth as cells transformed with recombinants that express wild-type (wt) LMP1 (7). Within the proximal 45 residues of the CT, residues 201–210 are the core of a domain that enables LMP1 to associate with tumor necrosis factor receptor associated factors (TRAFs; refs. 8–10). Because the effects of LMP1 on B lymphocyte growth are remarkably similar to those induced by CD40, a tumor necrosis factor receptor family member that also interacts with TRAFs, the association of LMP1 with TRAFs is likely to be important in LMP1’s effects on cell growth (1, 8, 11–17).
Figure 1.
Structure of LMP1. The Flag epitope was introduced at the LMP1 amino terminus (NH2). LMP1 residues 185, 211, 231, and 386 are noted. The single TRAF binding site and the two NF-κB-inducing domains are indicated.
The experiments described here use recombinant EBV genetic analyses to evaluate the specific importance of the LMP1 TRAF binding domain in B lymphocyte transformation. The findings that EBV recombinants expressing an LMP1 truncated after the first 45 residues of the CT can transform primary B lymphocytes, whereas recombinants expressing LMP1 truncated before the first 45 residues of the CT cannot, simply indicate that the 45 residues of the CT are sufficient for transformation. These findings do not exclude the possibility that the rest of the LMP1 CT could be a second domain that is sufficient for transformation in the absence of the first domain. In fact, LMP1 activation of NF-κB (18, 19) could be a key effector in B lymphocyte transformation and most of the NF-κB activating effects of LMP1 are mediated by the distal 35 residues of the CT (20, 21). The first 45 residues of the LMP1 CT transduce about 25% of the LMP1-mediated NF-kB activation, and this appears to be mediated by TRAF1 and TRAF2 heterodimers (9, 10). If NF-κB activation is the principal effector of transformation, deletion of the TRAF binding domain from LMP1 might have less of an effect on growth transformation than truncation after the first 45 residues of the CT.
MATERIALS AND METHODS
Cells.
P3HR-1 (22), IB4 (23), BJAB (24), LCL, and 293 cells were grown as described (6, 9).
DNA Clones.
EcoRI A, SalI E/C, and pSVNaeZ were as described (25–27). Plasmid pL Flag LMP1 DNA was made by replacing codons 2–4 of S-wt (6) with codons for the Flag epitope (Kodak) between the ClaI and XbaI sites, placing a NotI site at a HindIII site at nucleotide 166480 and a PacI site at a BglII site (nucleotide 169037). Cosmid Flag LMP1 DNA joins EcoRI (nucleotide 95239)–NotI (nucleotide 117614) from EcoRI B with NotI (nucleotide 166480)–SalI (nucleotide 643) from pL Flag LMP1 with SalI (nucleotide 643)–SnaBI (nucleotide 13219) from SnaBI B with pDVcosPENBSP vector DNA (28). Cosmids were cloned as described (28). Codons 185–211 were deleted by PCR with pL Flag LMP1 with ΔL3′ (5′-gcctatgacatggtaatgcctag-3′) and Δ185–211 (5′-taatctggatgggccatgaatctgactctaac-3′), digesting the DNA with BsaBI and CelII, and inserting the DNA into pL Flag LMP1 to make pL Flag LMP1Δ185–211. Cosmid Flag LMP1Δ185–211 DNA is a replacement of the NotI to PacI DNA of cosmid Flag LMP1 DNA with that from pL Flag LMP1Δ185–211. Expression vectors pSG5 Flag LMP1 and pSG5 Flag LMP1Δ185–211 are 2.4-kb MluI DNA from pL Flag LMP1 or pL Flag LMP1Δ185–211 inserted into pSG5 (Stratagene). GAL4–LMP1 fusions were expressed in pAS1-CYH2. GAL4 LMP1 182–386 and GAL4 LMP1Δ185–211 are BsaBI–BglII DNAs from pL Flag LMP1 and pL Flag LMP1Δ185–211.
Yeast Two-Hybrid Assay.
The methods of yeast culture, transformation, and β-galactosidase detection were as described (29).
NF-κB Activation.
293 cells (5 × 106) were electroporated with LMP1 expression vector, 3x-κB-L luciferase reporter or mut-κB-L (21) and pGK–β-gal transfection control and analyzed as described (9).
Recombinant EBV Construction.
Methods for making recombinant EBV from three cosmids were as described (28, 30).
PCR, Southern Blot, Western Blot, and in Situ Immunofluorescence Analyses.
The methods of cell and virus sample preparation and detection have been described (6, 7, 28).
RESULTS
LMP1Δ185–211 Does Not Interact with TRAF3 and Has Slightly Reduced NF-κB Activating Effects.
Recombinant EBV with a Flag epitope insertion in the LMP1 amino terminus are fully competent for transforming primary B lymphocytes into LCLs (9). The Flag epitope facilitates biochemical analyses because most LMP1 antibodies recognize the CT and interfere with efficient TRAF coprecipitation. Flag LMP1 DNA was further mutated by in-frame deletion of codons 185–211 (Fig. 1). Cotransfection of Flag LMP1 or Flag LMP1Δ185–211 DNAs with EBNA 2 expression vector DNA into EBV-negative B lymphoma cells resulted in 60- or 57-kDa proteins recognized on immunoblots by M5 antibody to the Flag epitope or by S12 (31) monoclonal antibody to the LMP1 CT (data not shown).
TRAF1, TRAF2, and TRAF3 bind to a single site at residues 199–214 in the LMP1 CT (9, 10). TRAF3 binds most strongly, and TRAF2 binds least strongly. To confirm that deletion of residues 185–211 has removed the only TRAF binding site, LMP1Δ185–211 and wt LMP1 were tested for TRAF3 interaction by yeast two-hybrid assay (8). LMP1 fused to the GAL4 DNA binding domain was scored for interaction with TRAF3 residues 314–562 fused to the GAL4 transactivating domain by activation of a β-galactosidase reporter. GAL4–LMP1 residues 182–386 strongly interacted with TRAF3, whereas GAL4–LMP1 182–386 deleted of residues 185–211 did not interact with TRAF3. These studies in yeast and those in lymphocytes described below indicate that LMP1Δ185–211 does not directly associate with TRAFs.
LMP1Δ185–211 was also tested for similarity to wt LMP1 in NF-κB activation. As expected from previous reports, the distal LMP1 CT effected about 75% of wt LMP1 NF-κB activation (9, 20, 21). In 293 cells, pSG5 Flag LMP1 activates a luciferase reporter with three class 1 major histocompatibility complex-derived NF-κB sites and a minimal fos promoter (21) 28-fold (SEM ± 2.2) compared with pSG5 vector control, whereas pSG5 Flag LMP1Δ185–211 activates 20 ± 2.0-fold. NF-κB inductions by pSG5 Flag LMP1 1–231 (3.5 ± 0.1-fold) or pSG5 Flag LMP1 1–351 (3.2 ± 0.1-fold) were somewhat less than the 10-fold level expected (9, 20, 21). Immunoblots probed with M5 antibody to the Flag epitope (data not shown) indicated that mutant and wt LMP1 expression levels were similar in the transfected 293 cells.
Recombinant EBV with Flag LMP1 and Flag LMP1Δ185–211 Genes.
EBV recombinants with Flag LMP1 or Flag LMP1Δ185–211 genes were generated by replacement cloning of pL Flag LMP1 or pL Flag LMP1Δ185–211 DNAs into their natural site in an EBV cosmid and by homologous recombination with two other overlapping EBV cosmids in cotransfected P3HR-1 cells (28). This three-cosmid recombination method was used because of the predicted higher efficiency of incorporation of the cosmid that contains mutated LMP1 DNA into EBV recombinants (28). P3HR-1 cells are infected with a replication-competent EBV that provides replication and packaging functions to the transfected, overlapping cosmid DNAs. The P3HR-1 EBV DNA replicates and is packaged into infectious virions, but it cannot transform B lymphocytes due to a deletion that includes EBNA 2 DNA and part of EBNA LP DNA (32–34). The replicating P3HR-1 EBV DNA can also recombine with the cosmids to generate various recombinants, including wt transforming EBV (28).
The three overlapping cosmids were electroporated into P3HR-1 cells, virus replication was induced, the resultant virus was used to infect primary B lymphocytes, and the infected cells were cultured in microtiter plates under conditions where there was less than one transformed cell per well (28). About 10,000-fold more nonrecombinant P3HR-1 virus is produced than transforming recombinants in such experiments, and more than half of the LCLs that grow out are coinfected with P3HR-1 EBV (5–7, 25–28). P3HR-1 EBV expresses wt LMP1 that can complement the mutated LMP1 encoded by an EBV recombinant that has wt EBNA 2 and EBNA LP genes. Over the first several weeks in culture, cells infected with a transformation-competent EBV recombinant grow out as LCLs, and all other cells die. In previous experiments, more than one-third of the transforming EBV recombinants were found to be composed primarily or entirely of the transfected cosmid DNAs. The rest were composed of cosmid DNAs that had recombined with P3HR-1 DNA. Overall, about half of the EBV recombinants had DNA from the transfected LMP1 cosmid (28, 35).
The resultant LCLs were scored for the Flag mutation by PCRs with primers that amplify a Flag-specific 275-bp DNA from Flag LMP1 or Flag LMP1Δ185–211 DNA and a 257-bp DNA from wt P3HR-1 LMP1 DNA. Primers that amplify a 459-bp DNA from Flag or wt LMP1 DNA and a deletion-specific 378-bp DNA from Flag LMP1Δ185–211 DNA were also used. Flag LMP1 genes were detected in only 7 of 45 LCLs (16%) arising from infection of human B lymphocytes with virus from P3HR-1 cells transfected with cosmid Flag LMP1 DNA. One of these seven LCLs (F-L1) was infected with only Flag LMP1 EBV DNA and lacked wt LMP1 DNA from a coinfecting P3HR-1 EBV. The six remaining LCLs, F-L2 to F-L7, were coinfected with P3HR-1 EBV, and five of these remained coinfected during subsequent cultivation. All of the F-L cell lines were easily propagated to high cell numbers, confirming that the Flag epitope mutation does not adversely affect cell growth. In comparison, Flag LMP1Δ185–211 was detected in 8 of 90 LCLs (9%) arising from infection of B lymphocytes with virus from P3HR-1 cells transfected with cosmid Flag LMP1Δ185–211 DNA. These LCLs, F-Δ1 to F-Δ8, have Flag LMP1Δ185–211 and wt LMP1 from coinfecting P3HR-1. Five of these LCLs, designated F-ΔL1 to F-ΔL5, were successfully propagated to high cell numbers for further study, whereas three others could not be maintained in long-term cultivation.
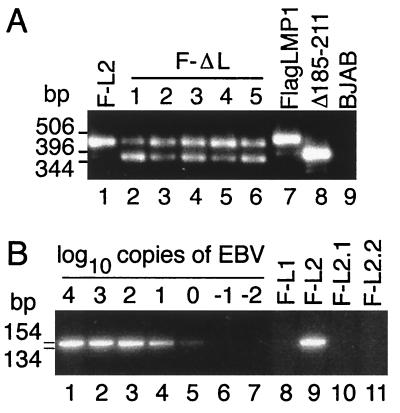
To evaluate the growth-transforming potential of recombinants with the Flag LMP1Δ185–211 gene versus Flag LMP1 controls, virus replication was induced in the coinfected LCLs, and the virus progeny was used to infect primary B lymphocytes in a clonal transformation assay. In Fig. 2A, the presence of recombinant and P3HR-1 genomes in 0.45-μm filtered supernatants from LCLs that had been induced to lytic EBV infection was confirmed by PCR. In lanes 2–6 of Fig. 2, equimolar amounts of a 459-bp fragment indicative of wt LMP1 and a 378-bp fragment indicative of Flag LMP1Δ185–211 DNA were amplified from these virus preparations, demonstrating that wt and Flag LMP1Δ185–211 DNAs were packaged into virions with similar efficiency. A 378-bp DNA is amplified from control pL Flag LMP1Δ185–211 DNA (Fig. 2, lane 8), whereas a 459-bp DNA is amplified from control wt LMP1 DNA (Fig. 2, lane 7) and from lytically infected LCL F-L2, which is coinfected with P3HR-1 and Flag LMP1 recombinant EBV (Fig. 2, lane 1). As expected, supernatants from EBV-negative BJAB cells or latently infected IB4 had neither wt nor Flag LMP1Δ185–211 DNA (Fig. 2, lane 9, and data not shown).
Figure 2.
PCR analysis of LMP1 DNA. (A) LMP1 DNAs in supernatants from lytically infected LCLs coinfected with P3HR-1 and Flag LMP1 recombinant EBV (F-L2 in lane 1) or P3HR-1 and Flag LMP1Δ185–211 recombinant EBV (F-ΔL1 to F-ΔL5 in lanes 2–6) were analyzed by PCR with ΔL5′ (5′-ctctattggttgatctcctttgg-3′) and 3′L315 (5′-attgtggagggcctccatcatttc-3′). Control DNAs pL Flag LMP1 (lane 7) and pL Flag LMP1Δ185–211 (lane 8), and DNA from EBV-negative BJAB cells (lane 9) were also analyzed. Size standards in base pairs are noted to the left. (B). PCR detection of wt LMP1 DNA with 5′L1 (5′-cacgcgttactctgacgtagccg-3′) and WTND (5′-tcctcgagggggccgtcgc-3′). Lanes 1–7 contain 104 IB4 cells (4 genomes per cell) serially 10-fold diluted with 104 EBV-negative BJAB cells. The endpoint lies after lane 5 for a sensitivity of 4 wt LMP1 DNAs per 104 cells. wt LMP1 DNA is in LCL F-L2 (lane 9) but not in LCL F-L1 (lane 8) or LCLs F-L2.1 and F-L2.2 (lanes 10 and 11). Size standards in base pairs are to the left.
The results of PCR analysis for LMP1 genotype in the second generation LCLs that arose after infection with virus from the Flag LMP1Δ185–211 and P3HR-1 LMP1 coinfected LCLs are presented in Table 1. Under clonal conditions where fewer than 33% of the wells have a transformed cell, Flag LMP1Δ185–211 recombinants were not detected in these second-generation LCLs. The LCLs had only a wt P3HR-1 LMP1 gene as the result of secondary recombination between the coinfecting P3HR-1 EBV genome and the Flag LMP1Δ185–211 EBV recombinant genome that had been maintained in the original LCLs because it had the transformation-essential EBNA 2 and EBNA LP genes. The equal presence of Flag LMP1Δ185–211 and wt LMP1 in virus supernatants and the absence of Flag LMP1Δ185–211 under clonal transformation conditions in the resultant LCLs are clear evidence that this LMP1 mutation has substantially reduced or null transformation potential.
Table 1.
PCR analysis for LMP1 DNA in second-generation LCLs
| LCL source | LMP1 genotypes | Volume of virus, ml | Wells with LCLs, % | No. of mutants alone | No. of mutants and wt | No. of wt alone |
|---|---|---|---|---|---|---|
| F-ΔL1 | Δ185–211 + wt | 1.0 | 100 | 0 | 6 | 88 |
| Δ185–211 + wt | 0.6 | 27 | 0 | 0 | 124 | |
| F-ΔL2 | Δ185–211 + wt | 1.0 | 100 | 0 | 14 | 81 |
| Δ185–211 + wt | 0.1 | 31 | 0 | 0 | 70 | |
| Δ185–211 + wt | 0.01 | 3 | 0 | 0 | 5 | |
| F-ΔL3 | Δ185–211 + wt | 0.5 | 4 | 0 | 0 | 7 |
| F-ΔL4 | Δ185–211 + wt | 0.5 | 13 | 0 | 0 | 24 |
| Δ185–211 + wt | 0.05 | 2 | 0 | 0 | 3 | |
| F-ΔL5 | Δ185–211 + wt | 0.5 | 5 | 0 | 0 | 10 |
| F-L2 | Flag LMP1 + wt | 0.5 | 3 | 2 | 0 | 4 |
LCLs were induced to lytic infection, and 5 × 106 primary mononuclear cells were infected with the indicated volume of virus and seeded into microtiter plates. LCLs that grew out were scored for LMP1Δ185–211 DNA, Flag DNA, and wt LMP1 DNA by PCR.
When more virus stock was used to infect primary B lymphocytes, nearly 100% of the microtiter wells had LCLs, and 6 of 94 LCLs transformed with virus from LCL FΔL-1 and 14 of 95 LCLs transformed with virus from LCL FΔL-2 had both wt LMP1 and Flag LMP1Δ185–211. This confirms that infectious Flag LMP1Δ185–211 EBV genomes are in the filtered virus preparations and can be found in newly transformed B lymphocytes when wt LMP1 is provided to the same cell by coinfection with P3HR-1 EBV that has wt LMP1. The absence of LCLs infected with an EBV genome with Flag LMP1Δ185–211 alone and the repeated finding of LCLs infected with EBV genomes produced by secondary recombination between P3HR-1 (wt LMP1 DNA but deleted for EBNA 2 and LP DNA) and Flag LMP1Δ185–211 recombinants (wt for EBNA 2 and EBNA LP DNA) indicate that LMP1 residues 185–211 are critical for primary B lymphocyte growth transformation. Virus stocks from five different Flag LMP1Δ185–211 and P3HR-1 EBV coinfected LCLs yielded 412 LCLs transformed by wt LMP1 only secondary recombinants and none by Flag LMP1Δ185–211 recombinants only.
In sharp contrast, passage of virus from F-L2, an LCL coinfected with P3HR-1 and Flag LMP1 recombinant, into primary B lymphocytes resulted (Table 1) in 2 of 6 LCLs that were infected with Flag LMP1 recombinant alone, whereas the rest were infected with secondary recombinants. In Fig. 2B, LCLs infected with Flag LMP1 recombinants were scored for the presence of wt LMP1 genes by PCR using wt LMP1-specific primers that detect 4 copies of wt LMP1 DNA per 10,000 cells (lane 5). LCL F-L1, which was isolated directly from P3HR-1 cells transfected with the three overlapping cosmids, has no wt LMP1 (Fig. 2, lane 8), indicating that this LCL is infected only with a Flag LMP1 recombinant, whereas LCL F-L2 has wt LMP1 DNA (Fig. 2, lane 9), indicating that this LCL is coinfected with P3HR-1 EBV. LCLs F-L2.1 and F-L2.2 were infected with virus from LCL F-L2 but have no wt LMP1 DNA (Fig. 2, lanes 10 and 11). Thus, Flag LMP1 from an LCL coinfected with Flag LMP1 recombinant EBV and P3HR-1 does segregate from P3HR-1 LMP1 in clonal transformation assays, indicating that Flag LMP1 recombinants are competent for transformation and Flag LMP1Δ185–211 recombinants are not.
Southern Blot, PCR, Western Blot, and Immunofluorescent Analyses of Coinfected LCLs.
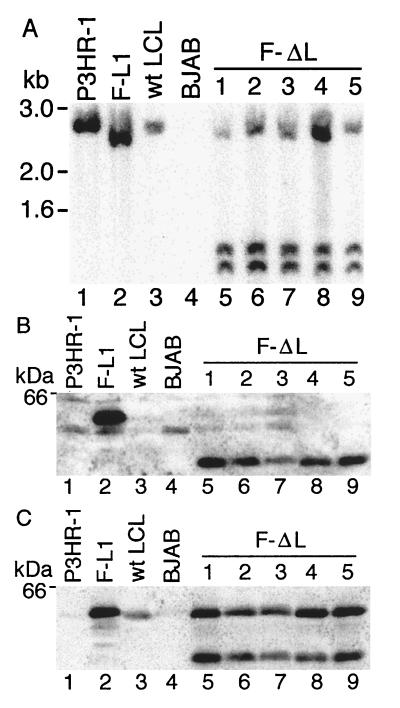
To evaluate whether genetic abnormalities or loss of Flag LMP1Δ185–211 expression might explain these results, LCLs F-ΔL1 to F-ΔL5, which are putatively coinfected with Flag LMP1Δ185–211 recombinant and P3HR-1 EBV, were examined by Southern blot, PCR, Western blot, and in situ immunofluorescence analyses. DNA extracted from LCLs was double-digested with MluI and SacI, size-separated, blotted to nylon filter, and probed with pL Flag LMP1 DNA. As shown in Fig. 3A, the probe hybridizes a 2.4-kb DNA from P3HR-1 (lane 1) or an LCL with a wt LMP1 gene (lane 3) and a 2.3-kb DNA from F-L1 LCL (lane 2) because the Flag alteration in Flag LMP1 DNA is linked to a new SacI site 158 bp from one of the MluI sites. (The 158-bp SacI–MluI fragment that would hybridize to the probe ran off the gel.) As expected, the probe detects nothing in EBV-negative B lymphoma cell line BJAB (Fig. 3, lane 4). In lanes 5–9 of Fig. 3, LCLs F-ΔL1 to F-ΔL5 have a dual pattern of hybridized DNA, demonstrating that they are coinfected with P3HR-1 (2.4-kb DNA) and a Flag LMP1Δ185–211 recombinant (1.1- and 1.2-kb DNAs due to a second SacI site after the end of the LMP1 coding sequence). These results demonstrate that the Flag LMP1Δ185–211 recombinant and P3HR-1 EBV-coinfected LCLs have LMP1 DNAs of the expected size and that these DNAs are present in equimolar amounts.
Figure 3.
Southern and Western blot analyses. (A) DNA from LCLs was cut with SacI and MluI, size-separated, and probed by Southern blotting with an EBV MluI DNA (nucleotides 167,129–169,560) which comprises LMP1 DNA. A 2.4-kb band from P3HR-1 (lane 1) and a wt EBV transformed LCL (lane 3) is recognized by this probe. In LCL F-L1 (lane 2), a SacI site near the Flag codons results in a 2.3-kb DNA, whereas the 0.16-kb band ran off the gel. In coinfected LCLs F-ΔL1 to F-ΔL5 (lanes 5–9), Flag LMP1Δ185–211 DNAs have a second SacI site near the last LMP1 codon, resulting in 1.2- and 1.1-kb DNAs, while the 0.16-kb DNA ran off the gel. The 2.4-kb DNA in lanes 5–9 is wt LMP1 DNA. DNA standards are noted on the left. (B) Denatured proteins from 5 × 104 LCL cells were size-separated, blotted to filters, and probed with M5 Flag antibody (Kodak). No signal is detected in P3HR-1 cells (lane 1), in a wt EBV-transformed LCL (lane 3), or in EBV-negative BJAB cells (lane 4). Flag LMP1 (60 kDa) is detected in LCL F-L1, whereas Flag LMP1Δ185–211 (57 kDa) is detected in LCLs F-ΔL1 to F-ΔL5 (lanes 5–9). A protein standard is noted on the left. (C) The same blot was stripped of antibody and reprobed with S12 monoclonal antibody to LMP1. All cells except EBV-negative BJAB (lane 4) demonstrate 60-kDa LMP1 or Flag LMP1.
PCR analysis performed on FΔL1 to FΔL5 and F-L1 LCLs with a primer specific for the terminal repeat and adjacent unique sequence DNA and a primer specific for the second exon in LMP1 DNA revealed the predicted amplified products of 880 bp from P3HR-1 and 908 bp from FΔL1 to FΔL5 and F-L1 LCLs. SacI digestion cleaved the 908-bp DNAs to 719 and 189 bp DNAs, which were clearly distinguishable in size from the SacI-resistant 880-bp P3HR-1 PCR-amplified DNA. These data confirm that wt P3HR-1 LMP1, Flag LMP1, and Flag LMP1Δ185–211 DNAs are each linked to the terminal repeat sequence in an EBV genome in these LCLs. Because the terminal repeat sequence contains the virion packaging signal, the Flag LMP1Δ185–211, Flag wt LMP1, and P3HR-1 LMP1 DNAs should each be packaged into virions during lytic infection in the respective coinfected cell lines.
Western blots of size-separated proteins from these LCLs were probed with M5 antibody to the Flag epitope. The results in Fig. 3B demonstrate that LCL F-L1 that is singly infected with a Flag LMP1 recombinant EBV expresses the expected 60-kDa protein (lane 2). The M5 antibody does not recognize a 60-kDa protein in P3HR-1 cells (lane 1), in a wt EBV-infected LCL (lane 3), or in an EBV-negative B lymphoma cell line (lane 4). LCLs F-ΔL1 to F-ΔL5 that are coinfected with Flag LMP1Δ185–211 recombinants and P3HR-1 have the expected M5-recognized Flag LMP1Δ185–211 protein of 57 kDa (lanes 5–9). When this blot was stripped of antibodies and then reprobed with S12 monoclonal antibody, which recognizes the LMP1 CT, 60-kDa wt LMP1 was also evident in all cells that have the 57-kDa Flag LMP1Δ185–211 (lanes 5–9). The low level of LMP1 in P3HR-1 cells (lane 1) is due to the absence of EBNA 2, which up-regulates LMP1 gene expression. These data indicate that the inability of the Flag LMP1Δ185–211-recombinant EBV to transform primary B lymphocytes is not due to lack of expressed protein.
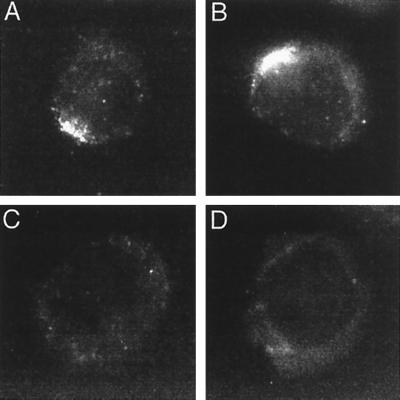
In Fig. 4, indirect immunofluorescent staining of cells with M5 antibody to the Flag epitope revealed specific recognition of Flag LMP1Δ185–211 or Flag LMP1 and no cross-reaction with LMP1 in P3HR-1 or in a wt EBV-infected LCL. Flag LMP1 (Fig. 4B) localized to aggregates in the plasma membrane and is similar to wt LMP1. Flag LMP1Δ185–211 (Fig. 4A) is expressed in the plasma membrane and also forms plasma membrane patches. Single gene transfer of Flag LMP1Δ185–211 expression vector DNA into EBV-negative B lymphoma revealed plasma membrane patching indistinguishable from LMP1 or Flag LMP1 (data not shown), indicating that the plasma membrane patching of Flag LMP1Δ185–211 does not require wt LMP1. Thus, the inability of Flag LMP1Δ185–211 recombinants to growth-transform cannot be attributed to improper localization.
Figure 4.
Immunofluorescent staining of cells with M5 antibody to the Flag epitope. (A) F-ΔL1, an LCL coinfected with P3HR-1 and Flag LMP1Δ185–211 EBV recombinant. (B) F-L1, an LCL infected with Flag LMP1 EBV recombinant only. (C) P3HR-1. (D) LCL infected with wt EBV.
Flag LMP1Δ185–211 Does Not Engage TRAF3 or TRAF1.
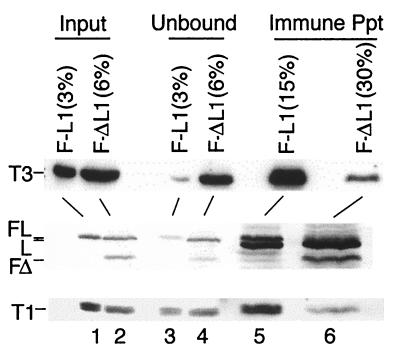
To investigate Flag LMP1Δ185–211 interaction with TRAFs in the coinfected LCLs, Flag LMP1Δ185–211 was immune-precipitated from LCL F-ΔL1, and Flag LMP1 was immune-precipitated from LCL F-L1 with M2 antibody to the Flag epitope. Coprecipitated proteins were scored by probing Western immunoblots with antiserum to TRAF3 (Fig. 5 Top), with S12 antibody to LMP1 (Middle), or with antiserum to TRAF1 (Bottom). The input amount of TRAF3, LMP1, and TRAF1 in unfractionated cell proteins from LCL F-L1 (lane 1) is similar to LCL F-ΔL1 (lane 2). The unbound TRAF3, wt LMP1, and TRAF1 that did not coprecipitate with M2 antibody was not significantly less than the input for LCL F-ΔL1 (lane 4), whereas TRAF3, Flag LMP1, and TRAF1 were depleted from LCL F-L1 lysates by immune precipitation (lane 3). TRAF3 and TRAF1 were easily detected in immune precipitates from LCL F-L1 (lane 5), whereas TRAF3 and TRAF1 were barely detectable in immune precipitates from LCL F-ΔL1 (lane 6). The amount of immune-precipitated Flag LMP1 (lane 5) is about the same as that of Flag LMP1Δ185–211 (lane 6). These results indicate that TRAF3 and TRAF1 do not associate with Flag LMP1Δ185–211 in vivo. LCL F-ΔL1 is coinfected with P3HR-1 and some wt LMP1 coprecipitated with Flag LMP1Δ185–211 (lane 6). The small amount of coprecipitated TRAF1 or TRAF3 is consistent with the small amount of coprecipitated wt LMP1.
Figure 5.
Flag LMP1Δ185–211 does not engage TRAF3 or TRAF1 in an LCL. F-L1 cells (107), an LCL infected with Flag LMP1 EBV recombinant, or 4 × 107 F-ΔL1 cells, an LCL coinfected with Flag LMP1Δ185–211 EBV recombinant and P3HR-1, were disrupted in Nonidet P-40 buffer. Part of the input cell lysate was withheld, and the rest was mixed with M2 antibody coupled to beads. Unbound proteins were separated from beads by centrifugation. Proteins coprecipitating with Flag LMP1 or Flag LMP1Δ185–211 were analyzed by Western blotting with anti-TRAF3 sera (Santa Cruz Biotechnology, top row), with S12 antibody to LMP1 (middle row), and with anti-TRAF1 sera (Santa Cruz Biotechnology, bottom row). Percentages indicate the fraction of the total cell lysate loaded into each lane. The positions of TRAF3 (T3), Flag LMP1 (FL), wt LMP1 (L), Flag LMP1Δ185–211 (FΔ), and TRAF1 (T1) are indicated to the left. Mouse Ig heavy chains (middle row, lanes 5 and 6) are just below Flag LMP1 and LMP1.
DISCUSSION
These experiments provide molecular genetic evidence that the TRAF binding domain of LMP1 is critical for primary B lymphocyte transformation. The evidence is based on the generation of infectious recombinant EBV genomes from overlapping cosmids in P3HR-1 cells which are infected with a replication-competent but transformation-negative EBV (32–34). Our analyses focused on Flag LMP1 or Flag LMP1Δ185–211 EBV recombinants that transformed primary B lymphocytes into LCLs alone or in cooperation with wt LMP1 provided by coinfection with P3HR-1. In these experiments, P3HR-1 EBV was produced in vast excess, and almost all LCLs infected with Flag LMP1 and all LCLs infected with Flag LMP1Δ185–211 containing genomes were also coinfected with a nonrecombinant P3HR-1 EBV genome. Thus, the only inference from these initial experiments was confirmatory evidence that Flag LMP1 recombinant EBV can transform primary B lymphocytes (9).
The derivation of five independent clones of Flag LMP1Δ185–211 LCLs that were each coinfected with P3HR-1 EBV enabled the induction of virus replication in these cells and further analyses of the outcome of transformation of primary B lymphocytes with the resultant EBV recombinants. LCLs singly infected with a Flag LMP1Δ185–211 EBV recombinant were never obtained, although more than 400 LCLs that had been infected with virus stocks that were nearly equimolar mixtures of Flag LMP1Δ185–211 and P3HR-1 wt LMP1 EBV genomes were analyzed. LCLs (412) were singly infected with a secondary recombinant that has only the P3HR-1 wt LMP1 gene. Twenty Flag LMP1Δ185–211-infected LCLs were obtained when more virus stock was used. Each Flag LMP1Δ185–211-infected LCL was coinfected with a wt LMP1 EBV genome. In sharp contrast, Flag LMP1 EBV recombinant segregated from P3HR-1 EBV in a parallel experiment. These data clearly indicate that residues 185–211 are critically required for LMP1’s role in primary B lymphocyte transformation.
The LMP1 185–211 sequence is likely critical for transformation because it has the single site through which LMP1 directly engages TRAFs (9, 10). The core of the TRAF binding site consists of residues 201–210. However, the efficiency with which this core binds TRAFs is less than that of slightly larger constructs, and negative effects of mutations within the core can be offset by wt sequence outside the core, indicating that surrounding residues are important in TRAF association. Furthermore, the last transmembrane domain ends near residue 185, leaving only about 16 residues before the core. Because almost all of LMP1 is stably associated with TRAF3, TRAF1, or TRAF2 in LCLs, residues surrounding the core are unlikely to be associated with other proteins. Thus, residues 185–211 are likely to be critically required because of their role in TRAF engagement. The failure of LMP1Δ185–211 to associate with TRAFs and to contribute to EBV-mediated growth transformation in the context of recombinant virus clearly implicates TRAF association in transformation and is fully consistent with the previous finding that LMP1 1–231 is sufficient for transformation and LCL outgrowth (7). Now that the TRAF binding core has been mapped to L201PHP QQATDD210 and mutations of P204 and Q206 have been shown to abrogate TRAF association (9), more precise recombinant EBV genetic analyses can be undertaken to further establish the biochemical and genetic linkage between TRAF association and transformation.
These results further implicating LMP1 interaction with TRAF as a critical component of EBV-mediated transformation are consistent with the similarity of LMP1 to CD40, CD30, or TNFRII in inducing lymphocyte growth, activation markers, adhesion molecules, and NF-κB (1, 8–19, 36–41). LMP1’s effects are most similar to those of CD40. LMP1 appears to activate NF-κB by primarily engaging TRAF1 and forming TRAF1 and TRAF2 heterodimers, whereas CD40 appears to directly engage TRAF2 (9, 37). Both LMP1 and CD40 also engage TRAF3, but TRAF3’s role in mediating LMP1 and CD40 effects other than as a negative modulator of NF-κB activation is uncertain (9, 37, 42).
The new finding that LMP1Δ185–211 EBV recombinants are defective in primary B lymphocyte transformation, despite nearly wt NF-κB activation, together with the previous findings that LMP1 1–231 is sufficient for primary B lymphocyte growth transformation, despite its ability to mediate only 25% of wt LMP1 NF-κB activation, indicate that NF-κB is not the dominant effector of transformation and that the 187–231 domain has transforming effects beyond those mediated by NF-κB activation. This is consistent with the recent finding that, whereas both LMP1 1–231 and LMP1Δ187–351 can activate the NF-κB-responsive A20 gene, only LMP1 1–231 can also up-regulate epidermal growth factor receptor expression in a colon carcinoma cell line (43). Further, TRAF1 and TRAF2 heterodimers have been implicated in interactions with IAPs, the putative inhibitors of apoptosis (44–46).
These experiments support the working hypothesis that further delineation of the role of TRAFs in LMP1, CD40, and CD30 effects on cell growth is important for the rational development of therapeutics for the control of EBV-associated malignancies. LMP1 is expressed at an early stage in EBV-associated lymphoid and epithelioid malignancies and in EBV-associated Hodgkin disease. Interestingly, initial nasopharyngeal carcinoma and Hodgkin disease tumors consist of mixtures of malignant cells and normal T lymphocytes. Hodgkin disease tumor cells express large amounts of CD30, and nasopharyngeal carcinoma cells frequently express CD40. The tumor cells cannot be grown in pure culture and may depend on T cell expression of CD30 and CD40 ligands and on adjuvant signaling from CD30 or CD40. Because of the similarity in LMP1, CD40, and CD30 interactions with TRAFs, inhibitors of TRAF signaling may have dual utility in containing the growth of these tumor cells (12–15, 38, 40, 41).
Acknowledgments
We thank Erle Robertson and George Mosialos for providing invaluable advice and reagents for this manuscript. This work was supported by Public Health Service Grants CA47006-06 (E.D.K.), CA67380-02 (K.M.K.), and CA01568-04 (K.M.K.) from the National Cancer Institute. K.M.I. is a postdoctoral fellow of The Medical Foundation—Charles A. King Trust.
Footnotes
Abbreviations: EBV, Epstein–Barr virus; LMP1, latent membrane protein 1; LCLs, lymphoblastoid cell lines; TRAF, tumor necrosis factor receptor associated factor; CT, carboxyl terminus; wt, wild type.
References
- 1.Rickinson A B, Kieff E. In: Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Philadelphia: Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Raab T N. Am J Pathol. 1995;146:1355–1367. [PMC free article] [PubMed] [Google Scholar]
- 3.Fennewald S, van Santen V, Kieff E. J Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liebowitz D, Wang D, Kieff E. J Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaye K M, Izumi K M, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumi K M, Kaye K M, Kieff E D. J Virol. 1994;68:4369–4376. doi: 10.1128/jvi.68.7.4369-4376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye K M, Izumi K M, Mosialos G, Kieff E. J Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 9.Devergne O, Hatzivassilliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieff E. In: Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Philadelphia: Raven; 1996. pp. 2343–2396. [Google Scholar]
- 12.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi J P, van Kooten C, Liu Y J, Rousset F, Saeland S. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 13.Kehry M R. J Immunol. 1996;156:2345–2348. [PubMed] [Google Scholar]
- 14.Noelle R J. Immunity. 1996;4:415–419. doi: 10.1016/s1074-7613(00)80408-2. [DOI] [PubMed] [Google Scholar]
- 15.Boise L H, Thompson C B. Science. 1996;274:67–68. doi: 10.1126/science.274.5284.67. [DOI] [PubMed] [Google Scholar]
- 16.Hu H M, O’Rourke K, Boguski M S, Dixit V M. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 17.Cheng G, Cleary A M, Ye Z S, Hong D I, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 18.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 19.Hammarskjold M L, Simurda M C. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huen D S, Henderson S A, Croom C D, Rowe M. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 21.Mitchell T, Sugden B. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G, Robinson J, Heston L, Lipman M. Proc Natl Acad Sci USA. 1974;71:4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menezes J, Liebold W, Klein G, Clements G. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 25.Tomkinson B, Kieff E. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomkinson B, Robertson E, Kieff E. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan S, Tomkinson B, Kieff E. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson E S, Tomkinson B, Kieff E. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durfee T, Becherer K, Chen P-L, Yeh S, Yang Y, Kilburn A E, Lee W, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 30.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann K P, Staunton D, Thorley-Lawson D. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen J I, Wang F, Mannick J, Kieff E. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammerschmidt W, Sugden B. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 34.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson E, Kieff E. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 37.Rothe M, Sarma V, Dixit V M, Goeddel D V. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 38.Ansieau S, Scheffrahn I, Mosialos G, Brand H, Duyster J, Kaye K, Harada J, Dougall B, Hubinger G, Kieff E, Herrmann F, Leutz A, Gruss H. Proc Natl Acad Sci USA. 1996;93:14053–14058. doi: 10.1073/pnas.93.24.14053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. J Virol. 1996;24:1879–1885. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S Y, Lee S Y, Kandala G, Liou M L, Liou H C, Choi Y. Proc Natl Acad Sci USA. 1996;93:9699–9703. doi: 10.1073/pnas.93.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gedrich R W, Gilfillan M C, Duckett C S, Van Dongen J L, Thompson C B. J Biol Chem. 1996;271:12852–12858. doi: 10.1074/jbc.271.22.12852. [DOI] [PubMed] [Google Scholar]
- 42.Cheng G, Baltimore D. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 43.Miller W, Mosialos G, Kieff E, Raab-Traub N. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothe M, Pan M G, Henzel W J, Ayres T M, Goeddel D V. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 45.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J E, MacKenzie A, Korneluck R G. Nature (London) 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 46.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]