Abstract
The two-component signal transduction pathways in bacteria use a histidine–aspartate phosphorelay circuit to mediate cellular changes in response to environmental stimuli. Here we describe a novel two-component todST system, which activates expression of the toluene degradation (tod) pathway in Pseudomonas putida F1. The todS gene is predicted to encode a sensory hybrid kinase with two unique properties—a basic region leucine zipper dimerization motif at the N terminus and a duplicated histidine kinase motif. Evidence from a synthetic peptide model suggests that TodS binds as a dimer to a pseudopalindromic sequence (5′-TGACTCA), which resembles the recognition sequence of the eukaryotic transcription factors Fos and Jun. These results provide additional evidence that bacteria and eukaryotes share common regulatory motifs. The todT gene product, a response regulator, was overproduced as a fusion protein in Escherichia coli, and the purified protein was found to bind specifically to a 6-bp palindromic DNA structure in the tod control region. The phosphorylated form of TodT appears to be the activator of tod structural genes. This is the first report of a two-component system that regulates aromatic metabolism in bacteria.
Keywords: two-component regulatory system, signal transduction, aromatic hydrocarbons, Pseudomonas
The toluene degradation (tod) pathway in Pseudomonas putida F1 (PpF1) is a paradigm of bacterial metabolism of aromatic hydrocarbons, which include the intractable polychlorinated biphenyls and some polynuclear aromatics, whereby ring fission is initiated through the formation of a “dihydrodiol” intermediate (1). The tod pathway is distinct from four other aerobic toluene-degrading routes of which regulation of the respective pathway falls into one of three existing families of aromatic catabolic regulators: the LysR transcriptional regulators, the σ54-dependent NtrC transcriptional activators, and the AraC/XylS activators (2). Although much is known about the biochemical steps and enzymology of the tod system there has been a paucity of information on its regulation (1, 3).
The two-component signal transduction systems are the major routes bacteria use to detect environmental signals that mediate changes in cellular behavior or biological processes (4, 5). These systems consist typically of two proteins—a sensory histidine kinase and a response regulator. In the general model, signal perception by the sensor, either membrane-bound or cytoplasmic, promotes autophosphorylation of a conserved histidine residue usually situated at the C terminus of the protein. The phosphate is then transferred from histidine to a conserved aspartate residue, usually found at the N terminus of the cognate response regulator. The phosphorylated response regulator subsequently mediates control at the transcriptional level by binding to a target DNA site(s). To date, several subtypes of the sensory component exist; a common variant are those known as “hybrid kinases,” which have both a histidine-kinase domain and a response-regulator domain in the same protein (for review see ref. 6). In addition, many of the hybrid kinases have a separate (extrinsic) response regulator. An example is the ArcAB system of Escherichia coli, which responds to osmolarity and redox changes. The kinase (ArcB) autophosphorylates at both the conserved histidine in the kinase domain and aspartate in the intrinsic response regulator domain. Phosphorylation of the extrinsic response regulator (ArcA) occurs from the conserved histidine and is dependent on phosphorylation of the ArcB intrinsic-response domain (7).
In this study we examined the PpF1 chromosomal region downstream of todH, the last gene of the tod operon (Fig. 1). Two regulatory genes, designated todS and todT, were cloned and their functional properties examined. These genes comprise a two-component system that provides a new and interesting insight into the regulation and activation of bacterial aromatic degradative pathways and an unique example of a basic region leucine zipper (bZIP) histidine kinase.
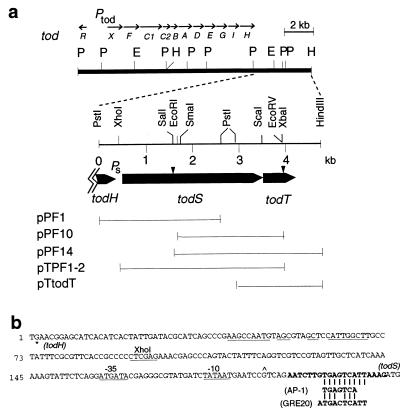
Figure 1.
(a) Localization of todS and todT genes in PpF1 chromosome. The todXFC1C2BADEGIH operon encodes a membrane protein (TodX) and seven enzymes (TodABC1C2, TodD–TodI) for the total conversion of toluene to pyruvate and acetyl-CoA (8, 9). todR is a truncated LysR-type protein and has no apparent regulatory role in tod expression (9). E, EcoRI; P, PstI; H, HindIII. Ptod and Ps are promoter sites. Sites of kanamycin (Km) resistance gene disruptions are indicated (arrowheads). Plasmid derivatives in the todST region are described in the text. (b) Characteristics of the todHS noncoding region. ∗, stop codon of todH. An inverted repeat sequence is underlined. Promoter elements resembling the E. coli −10 (TATAAT) and −35 (TTGACA) sequences, and Pseudomonas rpoD-like consensus sequences are underlined. A transcriptional start site (∧) was determined in E. coli by primer extension analysis (data not shown). The 20-mer sequence (in boldface type) in its double-stranded form was used in the gel retardation assays as described in Fig. 5. The AP-1-binding sequence of Jun/Fos (10) and yeast GCN4 transcription factors (GRE20; ref. 11) are provided for comparison.
MATERIALS AND METHODS
Localization and Inactivation of todST Genes.
Plasmid pDTG552 (12) was used as a probe to obtain three subsequent overlapping clones, pPF1 (2.7-kb PstI fragment), pPF10 (2.4-kb EcoRI–XbaI), and pPF14 (3.1-kb SalI–HindIII fragment), which encompass the todS and todT genes (Fig. 1). The vectors for the three plasmids are pK194 (13), pUC18, and pUC13 (14), respectively. Strain PpF1(todS::KmR) was constructed by first inserting a 2.2-kb SalI–XhoI fragment containing the kanamycin resistance (KmR) gene from Tn5 (15) into the SalI site of pPF1. An ≈0.3-kb SalI–SmaI DNA fragment containing the origin of transfer (oriT) of plasmid RK2 was inserted into the BamHI site of pPF1, which was then introduced into PpF1 by conjugation (9). Of 50 transconjugants analyzed, about one-half were the result of double crossovers. The double crossovers were detected by loss of the plasmid-encoded ApR and this result was subsequently supported by Southern hybridization (not shown). For inactivation of todT, a KmR cassette from pUC4K (Pharmacia) was inserted into the unique EcoRV site in todT gene in pPF14. The DNA was then introduced into PpF1 by conjugation. Transconjugants were designated PpF1(todT::KmR). A subclone of todT as a 0.66-kb PCR fragment inserted at the HincII site in pUC8 was named pUCtodT.
Overexpression of TodT and DNA Binding.
TodT was overproduced as a fusion protein using a commercial glutathione S-transferase (GST) system. A 630-bp NdeI–HindIII fragment containing todT in pUCtodT was gel purified and inserted into pGEX-4T-3 (Pharmacia) at the BamHI site by blunt-end ligation. The recombinant plasmid, which produced an in-frame TodT fusion to GST was designated pGSTtodT. A 49-kDa fusion protein was produced in E. coli XL-1 Blue cells induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside for 3 h. The fusion protein was purified according to the manufacturer’s instructions.
DNA binding was carried out as follows: primers merRX (5′-GAGCACGCACAGTATTAC) and merRP (5′-CATCGACATCAGGTTATC) were used to amplify the intergenic region of todRX with pRX2 (9) as the template. The gel-purified 253-bp PCR fragment was mixed in a 5:1 ratio with a 196-bp PCR DNA containing the todHS intergenic region (Fig. 1). Each tube containing 1 μg of DNA and varying amounts of protein (0, 1, or 3 μg) in 15 μl of binding buffer (20 mM Tris·HCl, pH 7.4/2 mM MgCl2/2 mM EDTA/10 mM KCl/0.1% Nonidet P-40) was incubated on ice for 20 min. Three microliters of gel-loading buffer was added and the products separated on a 4.5% polyacrylamide gel in TAE buffer (40 mM Tris·acetate/1 mM EDTA, pH 8.0) at 12°C (see Fig. 4a).
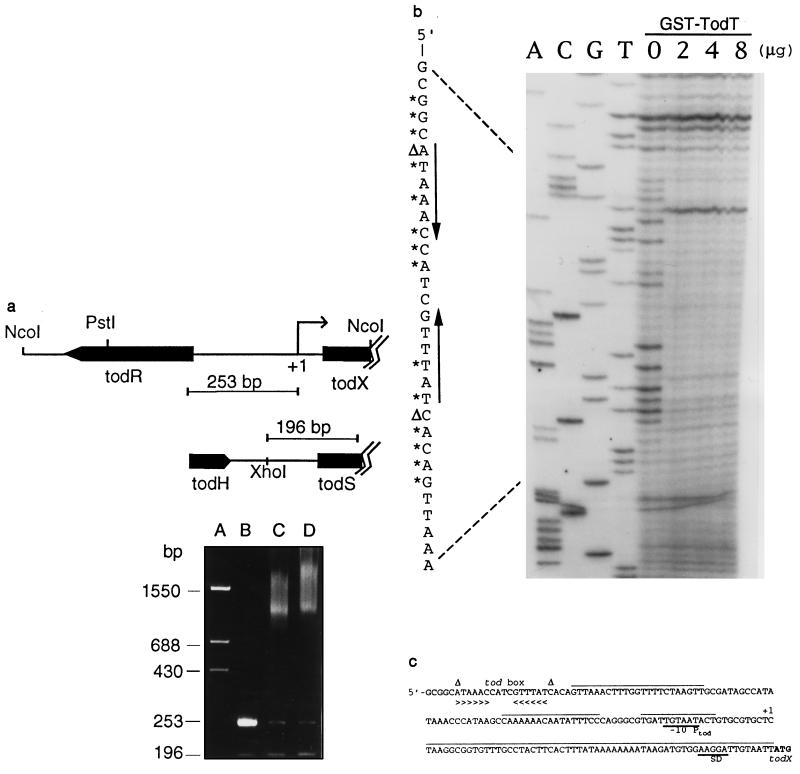
Figure 4.
(a) Specific binding of TodT to todRX intergenic region. DNA fragments (253 bp and 196 bp) used in gel retardation assays (lanes B–D) are as marked. Lanes: A, DNA standard; B, no protein; C, 1 μg protein; D, 3 μg protein (see Materials and Methods). (b) DNase I footprint of TodT binding at the tod promoter. The complement of the sequence surrounding the protected region is written alongside; * and Δ indicate DNase I protected and hypersensitive sites, respectively; converging arrows indicate a 6-bp inverted repeat labeled as tod box in c, together with other sequence characteristics of the tod regulatory region. +1, transcription start site and −10 promoter element (Ptod) were as determined previously (4). SD, Shine–Dalgarno sequence; A+T rich sequence is overlined.
DNase I Footprinting.
Footprinting of TodT to the tod promoter was carried out by the DNase I protection assay using the 253-bp PCR template as described, except that the primer merRX was labeled with [γ-32P]ATP by T4 polynucleotide kinase. The probe was precipitated with 20 μg of sonicated herring sperm DNA as a carrier, and dissolved in 70 μl of TE buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA). Two micrograms of the labeled DNA was incubated with varying amounts of the GST–TodT protein for 20 min on ice in 40 μl of binding buffer. DNase I (0.1 unit) and CaCl2 (2.5 mM) were added to the mixture and incubation was carried out on ice for 100 s. The reaction was terminated by adding 25 μl of stop solution containing 1% SDS, 125 mM NaCl, 25 mM EDTA, and 25 μg/ml tRNA. The mixture was extracted with phenol chloroform, followed by precipitation with ethanol. The pellet was dissolved in 14 μl of loading dye solution (80% formamide/1 mM EDTA/0.1% bromophenol blue/0.1% xylene cyanol). After heating at 90°C for 3 min, the samples were separated by electrophoresis on a 7% denaturing polyacrylamide gel containing 8 M urea along with a DNA sequence ladder generated with the labeled merRX as primer and plasmid pRX2 as template.
Site-Specific Mutagenesis of todT.
pGSTtodT(D56N) was generated by mutagenesis of the Asp-56 codon of todT in pGSTtodT. Two complementary mutagenic primers (A1, 5′-TACGGACATTCAAAATTAGAC and A2, 5′-GTCTAATTTTGAATGTCCGTA) and two flanking primers (B, 5′-GGCTCCGGCATATGAGTGATCGG and C, 5′-GCCATGAAGAGCTCCGACTATTCCAGG) were synthesized. The underlined base is the mutation designed to give N56 (AAT) instead of D56 (GAT) in the todT sequence. Two PCR reactions using pGSTtodT as template were carried out separately with primers A1 and B and A2 and C by conventional methods using Pfu DNA polymerase. The two PCR products were purified from an agarose gel. The products were mixed and subjected to PCR for 2 cycles followed by 35 cycles after addition of the 2 flanking primers B and C. The amplified DNA was digested with NheI and AflII and cloned into NheI–AflII-digested pGSTtodT. The sequence of the cloned fragment and site of mutation was confirmed by DNA sequencing.
Synthesis of TodSbs Peptide.
A 45-residue peptide, TodSbs, (MSSLDRKKPQNRSKNNYYNISLKEKGSEELTSEEHARIIFDGGGC)NH2, was synthesized on an Applied Biosystems model 430A synthesizer and was cleaved from the resin by hydrofluoric acid using conventional Boc-chemistry strategy. An acetamidomethyl group was used to protect the cysteine side chain during synthesis and the hydrofluoric acid cleavage. The sequence is amino acid 1–42 of TodS plus a C-terminal linker Gly-Gly-Cys, which is designed to replace functionally the leucine zipper (11). The two cysteines at positions 21 and 32 were replaced by Ser (underlined), since in several bZIP sequences these amino acids were found to be interchangeable (16).
Peptide Dimerization.
The peptide (25 mg) was dissolved in 10 ml of 80% acetic acid. An iodine solution (20 mg in 2 ml of 80% acetic acid) was added dropwise to deprotect the acetamidomethyl group and oxidize the cysteine residues for dimerization. The reaction solution was incubated for 2 h at room temperature and then cooled to 0°C. Unreacted iodine was neutralized by adding 0.1 N Na2S2O3 until the iodine color disappeared. The solvent was evaporated under vacuum. The peptide was dissolved in 5% acetonitrile and 25% acetic acid, and purified by preparative HPLC (Vydac C4, 4.6 × 25 cm) using a linear gradient of 14–54% acetonitrile in 0.1% TFA (0.33% per min gradient, 33 ml/min flow rate). The purified product was lyophilized with >95% purity based on a profile of an analytical HPLC (Vydac C18, 0.46 × 25 cm column, 10–50% acetonitrile in 0.1% TFA, 1.0% per min gradient, 1.0 ml/min flow rate) at 215 nm. The peptide composition was confirmed with a Beckman model 6300 amino acid analyzer and a Sciex (Thornhill, ON, Canada) API III mass spectrometer. The peptide showed correct amino acid composition and molecular mass (Mr, 10,259.3 Da observed vs. 10,259.5 Da calculated).
RESULTS AND DISCUSSION
TodS and TodT Proteins Belong to the Family of Two-Component Regulatory Systems.
Two open reading frames (ORFs), transcribed in the same direction as the core tod structural operon, were identified downstream of todH, the last gene of the tod operon, which encodes a 4-hydroxy-2-oxovalerate that carries out the penultimate step of toluene degradation to pyruvate (Fig. 1). The 2934-bp ORF, designated todS, encodes a 978-residue protein with a Mr of 108,225. Translational coupling of a second ORF of 206 residues (Mr, 23,030), designated todT, is suggested by an overlap of its potential start codon with the todS stop in the configuration ATGA. The predicted full-length sizes of TodS and TodT were verified by expression of subclones pTPF1–2 and pTtodT, respectively (Fig. 1) in the T7 polymerase/promoter system and labeling with [35S]methionine (not shown).
Analysis of the predicted amino acid sequence of TodS indicates that it is a new member of the hybrid class of histidine kinases (Fig. 2). More interestingly, the N terminus of TodS contains a motif characteristic of bZIP, which is common among transcriptional factors in organisms as diverse as fungi, plants, and mammals, but rarely described in prokaryotes (17, 22). The bZIP motif typically consists of a region with several basic residues, which probably contacts DNA directly and an adjacent region containing a heptad repeat of leucine (the leucine zipper) that mediates dimerization. The hypothesis that TodS dimerizes through its bZIP motif and binds to a specific sequence was tested and this is described later.
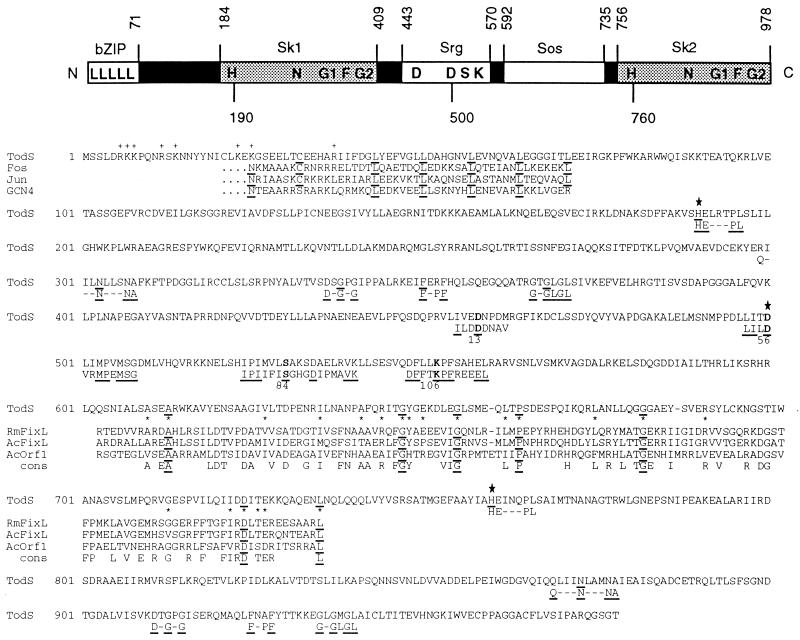
Figure 2.
(Upper) A schematic domain structure of TodS and its amino acid sequence characteristics. bZIP is characterized by the repeating heptad leucines (L) and the preceding charged (+) residues (17). (Lower) A comparison with eukaryotic Fos, Jun, and GCN4 bZIP sequences is shown; in TodS an invariant asparagine (N) found among eukaryotic bZIP proteins is replaced by a lysine (K), but a conserved cysteine (C) at position −11 from the first leucine is present. Sk1 and Sk2 are two histidine kinase domains characterized by the conserved amino acid blocks known as H (HE—PL), N (Q—N—NA), G1 (D-G-G), F (F-PF), and G2 (G-GLGL) as compiled by Parkinson and Kofoid (4). These conserved residues in TodS are underlined. Srg contains the conserved DDSK residues (in boldface type) characteristic of bacterial response regulators; the conserved Ser (S) is often replaced by a Thr (T) (18). Shown below the boldface residues, for comparison, are the corresponding regions from the TodT sequence. The large asterisks indicate potential phosphorylation sites. The Sos domain is defined by amino acid sequence similarity with the oxygen-sensing domain of Rhizobium meliloti FixL (19) and related proteins from Azorhizobium caulinodans (AcORF1) (20). Invariant residues are underlined; cons, consensus residues. The top three similarity scores as defined by the blast program (ref. 21; National Center for Biotechnology Information, Bethesda) for the Sk1 domain are: ORFX18 and PhoR of Bacillus subtilis (GenBank accession nos. L09228L09228 and M23549M23549, respectively); CpxA of E. coli [Protein Identification Resource (PIR) no. A29549A29549]; for Sk2: NodV and NswA of Bradyrhizobium japonicum (GenBank accession nos. M31765M31765 and Z22637Z22637, respectively); C4-dicarboxylate sensor kinase of Rhodobacter capsulatus (PIR no. S30288S30288); for Srg: PhoB of R. meliloti (GenBank accession no. M96261M96261), phosphate acceptor regulatory protein (CheY) of P. aeruginosa (GenBank accession no. X61231X61231), and PhoB of Klebsiella pneumoneae (GenBank accession no. M31794M31794).
Another novel feature of TodS is that it contains a duplicated histidine kinase domain, each of which is characterized by the five short sequence blocks, known as H, N, G1, F, and G2, that are highly conserved. Recently, the well-characterized ArcB and BarA sensory kinases regulating osmolarity in E. coli were reported to possess a second phosphorylatable histidine but lack the remaining conserved amino acid blocks (N, G1, G2, and F) in this second domain (23). The duplicated kinase regions (designated Sk1 and Sk2, spanning ≈aa 184–409 and ≈aa 756–978, respectively), have an overall 25% sequence similarity, which is a normal value when the sequences of different histidine kinase domains are compared. By analogy with known systems, His-190 and His-760 are the predicted autophosphorylated sites in TodS. Asp-458, Asp-500, Ser-530, and Lys-552 define the conserved residues in the “intrinsic” response regulator domain (designated Srg; spanning aa ≈443–570) of TodS. This ≈128-residue domain is reminiscent of CheY, which is a prototype response regulator of the bacterial chemotaxis pathway and of which the first three-dimensional structure of a response regulator has been determined (for review see ref. 18).
In between Srg and Sk2, spanning aa 592–735, is a putative oxygen-sensing region designated Sos. This assignment is based on its sequence similarity to a known oxygen-sensing/heme-binding domain of the R. meliloti FixL protein (19, 20). The low sequence homology was not unexpected, since heme-binding regions are known to be poorly conserved (19). The seven invariant amino acids as shown, in fact, exceed those of a previous alignment of sequences from two oxygenases, cytochrome P450 IID1 and isopenicillin synthase, which have similarities with the FixL heme-binding sequence (19).
Many histidine kinases contain a variable number of transmembrane domains along the polypeptide chains. TodS does not contain a region of sufficient hydrophobicity to suggest that it is a transmembrane protein (RAOARGOS and HELIXMEM analysis, PC/Gene, IntelliGenetics).
Analysis of the amino acid sequence of TodT reveals that it is most homologous (46% identity; 65% overall similarity) to the NodW response regulator of B. japonicum (24). Asp-13, Asp-56, Ser-84, and Lys-106 are the conserved amino acids typically found in response regulators (18). The C terminus of TodT contains a sequence (between aa 144 and 186 inclusive) that has 17 of 19 consensus residues of the DNA-binding domains of class 3 response regulators (25). These consensus residues are LSxREx2VLx5Gx2NKxIAx2Lx2Sx2TVx2Hx2Nx3KL, where the underlined sequences are conserved in TodT and x specifies other amino acids.
todST Inactivation and Positive Regulation by todT.
Several lines of evidence indicate that the todST gene products positively regulate expression of the tod structural genes. Both todS and todT genes were selectively interrupted at their chromosomal loci by insertion of a Kmr gene from Tn5 or pUC4K into the unique SalI and EcoRV sites, respectively, thus interrupting the TodSk1 domain and the putative DNA-binding region of TodT (Fig. 1). Mutants PpF1(todS::KmR) and PpF1(todT::KmR) resulting from double cross-over recombination were unable to use toluene as the sole carbon source. These strains also were incapable of converting indole to indigo, a reaction that is carried out by the toluene dioxygenase (TodABC1C2) complex (26). There also was no detectable catechol dioxygenase activity, indicating that the todE gene in these mutants was not expressed. To ascertain that the insertion of KmR in todS did not have a polar effect on the expression of todT, mutant PpF1(todS::KmR) was complemented with pAP13, a todS-containing plasmid in the broad-host range pVLT33 vector (27). The todS gene was carried on a 3.2-kb ScaI–ScaI fragment and cloned in the blunt-ended EcoRI site of pVLT33. As a result, PpF1(todS::KmR)[pAP13] was able to use toluene as a carbon source.
Further evidence for the regulatory role of todS and todT came from the absence of a toluene-inducible β-galactosidase (lacZ) activity when a reporter plasmid, pMR149, was mobilized into either mutant strain (Table 1). The pMR149 plasmid contains a toluene-inducible promoter element in front of the todX gene, which was transcriptionally fused to lacZ in the correct orientation to drive lacZ transcription (9). In toluene-grown PpF1 cells, pMR149 increased the lacZ activity nearly 5-fold, compared with no induction of the promoterless control plasmid pHRP311 (Table 1).
Table 1.
β-Galactosidase activities expressed from lacZ promoter fusion plasmids in PpF1 wild-type and mutant strains
| Strain | Plasmid | β-Galactosidase activities
|
||
|---|---|---|---|---|
| Uninduced | Induced | Fold induction | ||
| PpF1 (todS::KmR) | pMR149 | 240 | 300 | 1.0 |
| PpF1 (todT::KmR) | pMR149 | 370 | 470 | 1.2 |
| PpF1 | pMR149 | 350 | 1660 | 4.7 |
| PpF1 | pHRP311 | 250 | 260 | 1.0 |
Plasmid pHRP311 contains a promoterless lacZ gene (28) and was used as a negative control. Plasmid pMR149 has been described (9). Cultures were grown to midexponential phase in minimal salts medium with 20 mM pyruvate (uninduced) or 20 mM pyruvate plus toluene supplied in the vapor phase (induced). β-Galactosidase activity was assayed using chloroform and SDS to permeabilize cells and expressed in Miller units (9).
TodT is capable of activating the otherwise dormant tod operon in a heterologous system. E. coli HB101 cells containing plasmid pDTG301 were used as host. pDTG301 contains an intact todXFC1C2BADEGIH operon but lacks todT and the 3′ end of todS in the broad-host-range mobilizable pLARF1 vector (12). Despite the presence of the toluene dioxygenase-encoding genes (todC1C2BA) necessary for the indole–indigo conversion, cells of E. coli HB101(pDTG301) on Luria–Bertani plates were unable to carry out this reaction. When recombinant todT was provided in trans, indigo-forming colonies were obtained (Fig. 3). TodT (todTDN), in which Asp-56 has been changed to Asn-56 by site-specific mutagenesis, was unable to impart indigo production under the same assay conditions (Fig. 3b). The TodTDN variant, however, was still capable of specific DNA binding (not shown). Loss of function as a consequence of amino acid substitution at the conserved Asp (e.g., Asp-57 in CheY) of response regulators has been well documented (18). Consistent with the majority of response regulators (18), it appears that the phosphorylated form of TodT is the activator of the tod structural genes.
Figure 3.
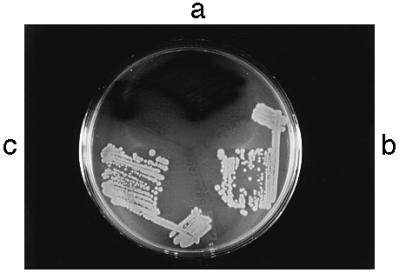
Positive transactivation action of TodT by indole assay and essentiality of Asp-56. The plasmids pGSTtodT (a), pGSTtodT(D56N) (b), and their parental vector pGEX-4T-3 (c) were transformed individually into E. coli HB101(pDTG301). A transformant from each was streaked on a Luria–Bertani plate containing 1 mM indole and the appropriate antibiotics. The plate was incubated overnight at 37°C, followed by an overnight incubation at room temperature. The strain containing wild-type TodT but not the mutant TodT in pGSTtodT(D56N) produced indigo colonies.
The trans-acting effect of TodT as a positive regulator of the tod operon in E. coli, in the absence of an intact todS, implies activation by metabolic cross-talk carried out either by a noncognate sensor or a chemical phosphorylating agent such as acetyl phosphate (29, 30). Alternatively, the truncated TodS (aa 1–375) encoded in pDTG301 is active in phosphorylation even though the predicted Sk1 domain, which extends to amino acid position 409 (Fig. 2) is incomplete.
Specific Binding of TodT to todX Promoter Region.
To study the DNA binding of TodT to the promoter region of todX, TodT was overexpressed as a GST fusion protein in E. coli, and purified using the glutathione-affinity chromatography (data not shown). Electrophoretic mobility-shift experiments were carried out using the purified protein and two PCR-generated DNA fragments; a 253-bp fragment upstream of the todX gene and a 196-bp fragment upstream of the todS gene as an internal control (Fig. 4a). A mixture of the two DNA fragments was used in the binding studies by incubating them with the purified TodT. As shown, TodT specifically shifted the 253-bp fragment and not the 190-bp fragment. In a separate experiment, specific DNA binding was also obtained with a 622-bp PstI–NcoI fragment containing the todRX intergenic region (data not shown). These results indicate that the TodT protein specifically binds to the todX promoter region.
To further localize the TodT-binding site, DNase I footprinting assays were performed using the 253-bp fragment labeled with 32P at one end (Fig. 4b). The studies showed that the TodT protected a small region encompassing a 6-bp inverted repeat (tod box) centered at −105 to −106 bp from the todX transcriptional start site (Fig. 4c). Two hypersensitive sites (marked ΔA and ΔC) are located at the base of the inverted repeat. These findings are consistent with the TodT being a positive regulator of the tod operon. One may also infer from the existence of the dyad symmetry that TodT binds to the protected region as a dimer.
AP-1 Binding.
The bZIP motif in transcriptional factors, such as the oncogene products Fos and Jun, and yeast GCN4 is known to mediate protein dimerization and bind DNA in a sequence-specific manner (10, 31). We asked whether this is the case for TodS, especially when the “pseudopalindromic” sequence, 5′-TGACTCA, identical to the recognition sequence of Fos and Jun (commonly known as the AP1-binding site; ref. 10), was uniquely found upstream of the todS start codon (Fig. 1). DNA-binding assays incorporated a short dimerized peptide provided by a Gly-Gly-Cys linker designed to mimic the full-length protein (11). Mobility-shift and competition assays indicate specific DNA-binding activity of the TodSbs dimerized peptide to two DNA substrates, each containing the AP-1 target sequence (Fig. 5). Binding to the 196-bp restriction fragment (Fig. 5b) was more pronounced than to the double-stranded 20-mer (Fig. 5a). In the latter case, although the binding appears weak (estimated to be 30%), molar excess of nonspecific poly(dI-dC) did not compete with the target DNAs and bovine serum albumin did not bind to the 20-mer. Reduction of the disulfide bond in TodSbs, by addition of 10 mM DTT, substantially decreased the amount of mobility-retarded DNA. This latter result lends support to the importance of dimerization for DNA binding, and provides the basis for further detailed investigations using purified TodS protein. TodS may be unique among known histidine kinases in incorporating a bZIP motif to effect protein dimerization for its function. Mutations that disrupt the bZIP motif in the full-length protein would test this hypothesis.
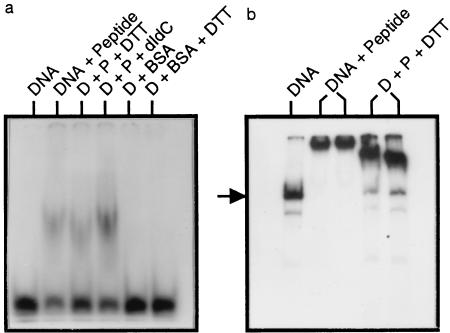
Figure 5.
Gel retardation assays showing DNA binding to a dimerized synthetic peptide. (a) The DNA substrate was a 32P-labeled double-stranded 20-mer, 5′-AATCTTGTGAGTCATTAAAG; the underlined portion is the eukaryotic AP-1 recognition sequence (see Fig. 1). P, peptide TodSbs; D, DNA samples; DTT, 10 mM dithiothreitol; BSA, bovine serum albumin; dIdC, poly(dI-dC) (Pharmacia). (b) The DNA substrate was a 32P-labeled 196-bp restriction fragment prepared by PCR amplification of the DNA region from an XhoI site in the todHS intergenic region to the nucleotides corresponding to the first leucine of the bZIP sequence. The arrow indicates the position of the 196-bp amplified DNA fragment. The minor faster migrating bands are unpurified PCR products. DNA binding was performed as described (6): 320 nM peptide/3 fmol of 20-mer/≈25 ng of PCR DNA (0.3 fmol). Binding was performed on ice for 30 min in buffer containing 20 mM Tris (pH 7.4), 2 mM MgCl2, 2 mM EDTA, 5 mM KCl, 0.1% Nonidet P-40. Mixtures were separated in 8% polyacrylamide gels in 0.5× Tris-borate EDTA buffer at 12°C.
Concluding Remarks.
We have described TodS, a novel histidine kinase that regulates toluene degradation in PpF1. The unusual features of TodS portray an ever-increasing diversity of the two-component systems in prokaryotes as well as those recently described homologues in plants and fungi (4–6). Indeed, much is to be learned about the prokaryotic devices for propagating dual, if not multiple, signals. The presence of a duplicated histidine kinase motif in TodS implies that it is likely a dual sensor responding to either toluene as a primary environmental signal or sensing oxidative stress in the cell milieu. Other possibilities exist. Because the tod pathway is oxygen-dependent, the presence of a putative oxygen-sensing domain in TodS may not be a fortuitous event. Because TodS is a cytoplasmic protein, an interaction(s) with a separate transmembrane (receptor) protein or another periplasmic protein(s) may be necessary. Preliminary evidence shows that TodX, an outer-membrane protein encoded by todX, the first gene of the tod operon, is somehow linked to the signal transduction process, which results in specific response of the tod promoter to toluene (Y.W. and P.C.K.L., unpublished work). We believe that greater specificity or fine tuning is bestowed upon catabolic promoter by the interplay of signal–transduction components in which transmission of the input and output signals have to be precise. This is in sharp contrast to promoters of other pseudomonad catabolic pathways in which a remarkable level of nonspecificity (leakiness) has been noted for the “single” transcriptional activators that control them. For example, the upper pathway promoter of the toluene/xylene (TOL) system in P. putida mt-2 can be efficiently cross-regulated by DmpR, the regulator of a different (phenol) catabolic pathway. Also, the toluene-responsive regulator of the TOL pathway (XylR), can recognize as effectors a whole variety of aromatic structures that are quite different from those substrates of the pathway (32).
It is evident that the basic principles of signal transduction involving protein phosphorylation in response to aromatic hydrocarbons are remarkably similar between toluene action in PpF1 and induction of cytochrome P450 A1 enzyme by dioxin (2,3,7,8-tetrachlorodibenzo-p-dioxin) in eukaryotes (for review see ref. 33). The existence of common regulatory features among bacteria and eukaryotes indicates that prokaryotes offer a rich ground for exploring and understanding key modes of cellular regulation, such as protein phosphorylation.
Acknowledgments
This is publication no. 40468 of the National Research Council of Canada. We thank M. Whiteway, D. Thomas, and C. Harwood for their valuable suggestions and comments, and J. Zimmermann for a critical reading of the manuscript. We appreciate the constructive comments of the anonymous reviewers and their suggestions on improving the manuscript. Ideas of this study stemmed from a sabbatical stay by P.C.K.L. in the laboratory of David T. Gibson (University of Iowa). M.R. was supported by U.S. Public Health Service Grant GM29909 to David T. Gibson from the National Institute of General Medical Sciences.
Footnotes
References
- 1.Zylstra G J, Gibson D T. In: Genetic Engineering: Principles and Methods. Setlow J K, editor. Vol. 13. New York: Plenum; 1991. pp. 183–203. [DOI] [PubMed] [Google Scholar]
- 2.van der Meer J R, De Vos W M, Harayama S, Zehnder A J B. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finette B A, Gibson D T. Biocatalysis. 1988;2:29–37. [Google Scholar]
- 4.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 5.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 6.Alex L A, Simon M I. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 7.Iuchi S. J Biol Chem. 1993;268:23972–23980. [PubMed] [Google Scholar]
- 8.Lau P C K, Bergeron H, Labbé D, Wang Y, Brousseau R, Gibson D T. Gene. 1994;146:7–13. doi: 10.1016/0378-1119(94)90827-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Rawlings M, Gibson D T, Labbé D, Bergeron H, Brousseau R, Lau P C K. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 10.Turner R, Tjian R. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- 11.Talanian R V, McKnight C J, Kim P S. Science. 1990;249:769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 12.Zylstra J, Gibson D T. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 13.Jobling M G, Holmes R K. Nucleic Acids Res. 1990;18:5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Lau P C K. Gene. 1996;168:15–21. doi: 10.1016/0378-1119(95)00732-6. [DOI] [PubMed] [Google Scholar]
- 16.O’Neil K T, Hoess R H, DeGrado W F. Science. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 17.Hurst C H. Protein Profile. 1994;1:123–168. [PubMed] [Google Scholar]
- 18.Volz K. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 19.Lois A F, Ditta G S, Helinski D R. J Bacteriol. 1993;175:1103–1109. doi: 10.1128/jb.175.4.1103-1109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahn D. Mol Microbiol. 1993;8:786–787. doi: 10.1111/j.1365-2958.1993.tb01623.x. [DOI] [PubMed] [Google Scholar]
- 21.Gish W, States D J. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 22.Kerppola T K, Curran T. Curr Opin Struct Biol. 1991;1:71–79. [Google Scholar]
- 23.Ishige K, Nagasawa S, Tokishita S, Mizuno T. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottfert M, Grob P, Hennecke H. Proc Natl Acad Sci USA. 1990;87:2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pao G M, Saier M H., Jr J Mol Evol. 1995;40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- 26.Ensley B D, Ratzkin B J, Osslund T D, Simon M J, Wackett L P, Gibson D T. Science. 1983;222:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 27.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Gene. 1993;123:17–23. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 28.Parales R E, Harwood C R. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 29.Wanner B L. J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCleary W R, Stock J B, Ninfa A J. J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentz R, Rauscher F J, III, Abate C, Curran T. Science. 1989;243:1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- 32.de Lorenzo V, Pérez-Martin J. Mol Microbiol. 1996;19:1177–1184. doi: 10.1111/j.1365-2958.1996.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 33.Whitlock J P., Jr Trends Endocrinol Metab. 1994;5:183–188. doi: 10.1016/1043-2760(94)90075-2. [DOI] [PubMed] [Google Scholar]