Abstract
The recently defined DEG/ENaC superfamily of sodium channels includes subunits of the amiloride-sensitive epithelial sodium channel (ENaC) of vertebrate colon, lung, kidney, and tongue, a molluscan FMRFamide-gated channel (FaNaC), and the nematode degenerins, which are suspected mechanosensory channels. We have identified two new members of this superfamily (BNaC1 and BNaC2) in a human brain cDNA library. Phylogenetic analysis indicates they are equally divergent from all other members of the DEG/ENaC superfamily and form a new branch or family. Human BNaC1 maps to 17q11.2-12 and hBNaC2 maps to 12q12. Northern blot and mouse brain in situ hybridizations indicate that both genes are coexpressed in most if not all brain neurons, although their patterns of expression vary slightly, and are expressed early in embryogenesis and throughout life. By analogy to the ENaCs and the degenerins, which form heteromultimeric channels, BNaC1 and BNaC2 may be subunits of the same channel.
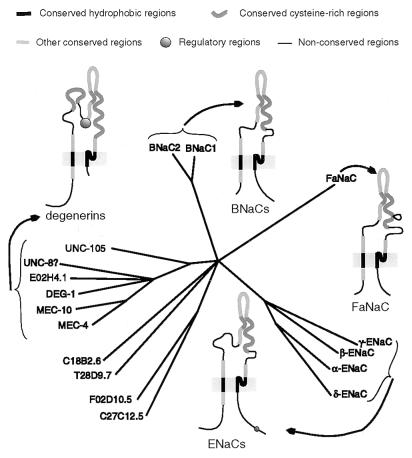
The recently defined DEG/ENaC superfamily of sodium channels contains to date 17 proteins (not counting the many orthologs found in different vertebrate species) that have similar sequences and the same predicted structure: intracellular N and C termini, two hydrophobic membrane-spanning regions, and a large extracellular loop, which contains many cysteine residues with conserved spacing. This topology has been experimentally demonstrated for members of two different branches: αENaC (1–3) and MEC-4 (4). In addition, all members that have been studied physiologically are selective for sodium and blocked by amiloride (5–13). Phylogenetic analysis of this superfamily reveals that it contains at least four branches or families (ref. 14; Fig. 1).
Figure 1.
Phylogenetic and structural comparison of DEG/ENaC superfamily members. The conserved hydrophobic regions (63 amino acids) of all these proteins were aligned, and the tree was generated by parsimony analysis using the paup program.
The ENaCs are expressed in the epithelia of the vertebrate kidney, colon, and lung, and are involved in sodium and water reabsorption in these tissues (5, 15, 16). Expression in tongue epithelium also suggests an indirect role in salty taste transduction (16). Three similar subunits (α, β, and γ-ENaC) form a heteromultimeric, constitutively-active channel when coexpressed in Xenopus oocytes (7). The reconstituted channel has properties nearly identical to that of the channel from epithelia, although biochemical experiments suggest that additional proteins form part of the native channel complex (17–19).
The degenerins of the nematode Caenorhabditis elegans are so named because rare, gain-of-function mutations in some of them cause swelling, vacuolation, and eventual death of some or all of the cells that express them (20–23). However, their normal function is thought to be mechanosensation, since loss-of-function mutations in two of them, mec-4 and mec-10, impair sensitivity to touch mediated by the six receptor neurons in which they are coexpressed (22, 24–26). It is believed that MEC-4 and MEC-10 are components of a mechanosensory channel complex, which would contain additional subunits or associated proteins. Although to date there are no electrophysiological recordings of any degenerin channel, a chimerical α-ENaC containing the predicted pore region of MEC-4, coexpressed with β-ENaC and γ-ENaC, results in an amiloride-sensitive sodium channel with distinct pore properties (10), suggesting that the degenerins are in fact channel proteins.
FaNaC, of the snail Helix aspersa, forms an amiloride-sensitive sodium channel that is activated by the peptide FMRFamide (9). This subunit is so far the only known member of that branch.
Here we define the BNaC family, an additional branch of the DEG/ENaC superfamily, composed of two genes (one previously characterized) that are expressed in brain.
MATERIALS AND METHODS
Cloning.
Unless otherwise indicated, all techniques were performed as described (27). Libraries were screened in aqueous media with probes obtained by random priming (28).
An 846-bp fragment contained within two expressed sequence tags (GenBank accession nos. Z45660Z45660 and F04549F04549) was obtained by PCR amplification from cDNA, which was synthesized from brain mRNA (CLONTECH): it contains codons 463–512 and some of the 3′ untranslated region (UTR) of hBNaC1. A partial cDNA clone (FCS16) of 1571 bp (codons 283–512 and the 3′ UTR) was obtained by screening a human frontal cortex cDNA library obtained from an 85-year-old female (Stratagene) and used as template to amplify a 639-bp fragment (codons 286–498), which was used to probe the library again to yield one slightly larger cDNA clone (codons 254–512 and the 3′ UTR). A 586-bp PCR fragment (codons 249–443) was used to probe another adult male human frontal cortex cDNA library (B616; ref. 29) at low stringency (final 3 washes at 40°C). One clone thus obtained (FC4-3) contained a complete coding sequence (CDS) for hBNaC1, 54 bp of the 5′ UTR, and the complete 3′ UTR. We also extended the partial cDNA sequence by 5′ RACE with primers AP1 (provided by CLONTECH) and 16A1 (5′-GGGTCTCACAGTCAATCCTACAGGCG-3′) or 16A2 (5′-GAAAGGTGGCTCAGACTGACTGTGGG-3′), and by 3′ RACE with 16S2 (5′-CCCACAGTCAGTCTGAGCCACCTTTC-3′) and AP1, using Marathon ready cDNA from a 37-year-old male as template (CLONTECH). The clones thus obtained contained larger 5′ UTR than clone FC4-3, but otherwise largely corresponded in sequence with it. The few nucleotide differences among the clones may be accounted for as PCR errors. The contig assembled with all of these clones corresponds to a 2748-bp cDNA (hBNaC1).
The human frontal cortex cDNA library hybridization also yielded a 1641-bp partial cDNA clone (FC3-1; containing 216 codons and a complete 3′ UTR) with a different 3′ UTR and a similar but not identical CDS than in hBNaC1. The library was probed again with a 947-bp EcoRI fragment from this clone (containing the 216 codons and part of the 3′ UTR). Of the three clones thus obtained, one of 2798 bp (FC6) contained a full CDS of 528 codons, 229 bp of predicted 5′ UTR, and 958 bp of 3′ UTR. Another partial cDNA clone (3007 bp; FC1-3) contained 138 additional bp of open reading frame between predicted codons 433 and 434 of clone FC6. The 138-bp segment was also found in some of the clones obtained by PCR amplification of brain cDNA from another individual. Therefore, the contigs assembled with all these clones correspond to alternative splice forms of hBNaC2, represented by 3785- and 3923-bp cDNAs (GenBank accession nos. U78181U78181 and U78180U78180, respectively).
Sequencing was performed by automated facilities at Massachusetts General Hospital and Harvard Medical School. Every segment of DNA was sequenced multiple times, and all coding regions were sequenced in both orientations. The nucleotide sequences of primers used in this study for PCR or for DNA sequencing are available upon request.
Northern Blotting.
Blots were purchased from CLONTECH and hybridized with probes obtained by random priming as suggested by the manufacturer. The human blots were hybridized with probes synthesized from the following templates: (i) 846 bp of hBNaC1 (codons 463–512 and 3′ UTR) obtained by PCR from one of the cDNA clones; (ii) the above described 947-bp EcoRI fragment obtained from hBNaC2 cDNA clone FC3-1. The mouse blot was hybridized with probes synthesized from the following templates: (i) 614 bp of mBNaC1 (GenBank accession no. U57353U57353; corresponding to codons 284–487 of hBNaC1), obtained by degenerate PCR from mouse brain cDNA; (ii) 597 bp of mBNaC2 (GenBank accession no. U78179U78179; corresponding to codons 330–528 of hBNaC2 shorter cDNA), similarly obtained. As a control, some blots were probed with human β-actin.
In Situ Hybridization.
A fragment of mBNaC1 corresponding to codons 284–437 of hBNaC1, and a fragment of mBNaC2 corresponding to codons 455–528 of the shorter hBNaC2 cDNA, were obtained by degenerate PCR using mouse brain cDNA as template. After subcloning into pCRII (Invitrogen) and sequencing the inserts, the clones were linearized and used as templates for in vitro transcription in the presence of digoxigenin-labeled UTP (Promega). To make the antisense riboprobe, the template was linearized with EcoRV and transcribed with SP6 RNA polymerase; to make the control sense riboprobe, the template was linearized with BamHI and transcribed with T7 RNA polymerase.
Mouse brain sagittal cryosections (14 micrometers thick) were fixed in 4% paraformaldehyde and hybridized overnight at 44°C (after 4 hr prehybridization) with the riboprobes in a previously described hybridization solution (28). After washes in 2× SSC (sodium chloride/sodium citrate; 10 min at room temperature), 2× SSC and 50% formamide (10 min at 50°C), 0.5× SSC and 50% formamide (10 min at 50°C), and 2× SSC (5 min at room temperature), the hybridized probes were detected with anti-digoxigenin antibodies linked to alkaline phosphatase (30). Nuclei were counterstained by incubating for 1 hr in 1 μM 4′,6-diamidino-2-phenylindole (DAPI).
Fluorescent in Situ Hybridization.
Fluorescent in situ hybridization on human chromosomes with biotinylated cDNA probes (31–33) was performed by SeeDNA (Toronto, Canada), using clone FCS16 of hBNaC1 and clone FC3-1 of hBNaC2. For each gene, hybridization to a pair of chromosomes was detected in 85% of mitotic figures (about 100 were observed), and all were in the same region. The map location was further defined by summarizing the results from 10 hybridizations.
RESULTS
In an effort to identify vertebrate homologs of the degenerin channels from C. elegans, we searched the data base of expressed sequence tags for sequences similar to various degenerins: deg-1 (23), mec-4 (4), mec-10 (22), and unc-105 (34). We found a human brain cDNA clone (c-zqg10; GenBank accession no. Z45660Z45660) with reasonable similarity to the second transmembrane domain of degenerins. By screening human brain cDNA libraries and by RACE we obtained an open reading frame for this gene, hBNaC1 (for human brain sodium channel 1), as well as of a close homolog, hBNaC2 (Fig. 2). In addition to several partial cDNA clones, we obtained two clones that lacked some of the 5′ or 3′ UTRs but nevertheless contained a CDS of either hBNaC1 or hBNaC2. The longest sequence [not counting the poly(A) tails] inferred for hBNaC1 contains 274 bp of 5′ UTR, 1536 bp of CDS, and 879 bp of 3′ UTR. For hBNaC2 the longest sequence contains 229 bp of 5′ UTR, 1584 or 1722 bp of CDS, and 1964 bp of 3′ UTR. Fragments of mouse homologs (mBNaC1 and mBNaC2) obtained by degenerate PCR were very similar to their human orthologs (99% identity at the amino acid level, in each case). The predicted sequences of hBNaC1 (512 amino acids) and hBNaC2 (528 amino acids for the shortest sequence encoded) were 74% identical over their first 465 residues, and diverged in sequence identity and length at their suspected intracellular COOH termini, only to regain conservation in the last eight amino acids, which form predicted sites for casein kinase II phosphorylation. Both sequences contained the hallmarks of the DEG/ENaC superfamily, including two hydrophobic stretches and, between them, regions with a conserved pattern of cysteines. However, both proteins lack the extracellular regions characteristic of the degenerin branch of this superfamily (refs. 14 and 23; Fig. 1).
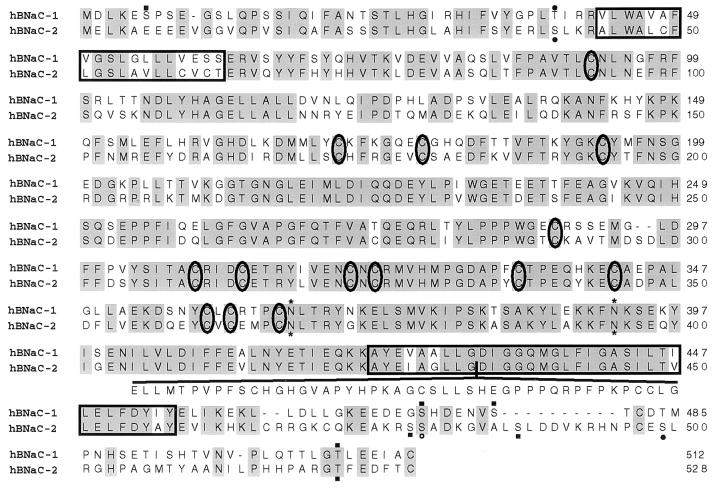
Figure 2.
Alignment of amino acid sequences predicted for human BNaC1 and BNaC2. Identical residues are shaded, hydrophobic regions are boxed, cysteines conserved in other DEG/ENaC proteins are circled, predicted N-glycosylation sites are indicated by asterisks, casein kinase II phosphorylation sites by solid squares, protein kinase sites by solid circles, and a cAMP- and cGMP-dependent protein kinase site by an open circle. The additional 46 amino acids predicted from one of the hBNaC2 cDNAs are indicated under the rest of the sequence. The amino acid sequence of hBNaC1 matches exactly that reported as mDEG (13), but differs from that reported as BNC1 (12), which contains an alanine rather than a threonine at position 495.
Some of the hBNaC2 clones, originating from the brain cDNA of different individuals by library screening or PCR amplification, contained an insert of 138 bp in the region encoding the second hydrophobic domain. This insert may correspond to the longer form of an alternatively spliced exon, because it contains a predicted splice donor site at its 5′ end, but no splice acceptor site at its 3′ end. The additional fragment of 46 amino acids is not hydrophobic, even though it is inserted in the second hydrophobic region, close to or at the pore. Computer analysis of this additional fragment by the methods of both Garnier–Robson (35) and Chou–Fasman (36) predict a secondary structure with several turns and coils.
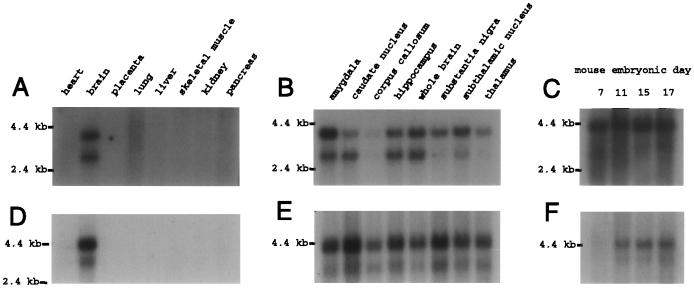
Northern blot hybridization (Fig. 3) revealed two bands for each gene, of roughly 2.8 kb and 3.8 kb for hBNaC1 and 3 kb and 4 kb for hBNaC2. The human cDNA sequences reported here are of the expected length for the lower mRNA band of hBNaC1 (2749 bp) and the higher mRNA band of hBNaC2 (3785–3923 bp). Both genes are expressed in brain but were not detected in heart, placenta, lung, liver, skeletal muscle, kidney, and pancreas (Fig. 3 A and D). All brain parts that we tested expressed at least the largest transcript of each gene, with one exception: the level of hBNaC1 in corpus callosum is nearly negligible, and the level of hBNaC2 in corpus callosum is less than elsewhere in the brain. Because in situ hybridization in mouse brain reveals no expression of either gene in corpus callosum, the Northern band may be contaminated with mRNA from a nearby brain area with higher expression of BNaC2 than BNaC1, such as choroid plexus (see below). To a limited extent transcripts are differentially expressed: the shorter hBNaC1 mRNA is abundant in amygdala, caudate nucleus, and hippocampus, whereas both hBNaC2 mRNAs are most abundant in caudate nucleus and substantia nigra (Fig. 3 B and E).
Figure 3.
Northern blot hybridization of human (A, B, D, and E) and mouse (C and F) BNaC1 (A–C) and BNaC2 (D–F). The blots contain mRNA from various human organs (A and D), parts of the human brain (B and E), or mouse embryos at several stages (C and F). Control hybridization of human β-actin cDNA to each blot (not shown) gave bands of nearly identical intensities in every lane, indicating that the amount of total mRNA per lane is about the same.
Northern blot hybridizations also indicated that the young mouse embryos express two transcripts of each ortholog: mBNaC1 mRNA is already present on day 7 of embryogenesis, and mBNaC2 mRNA is abundant by day 11 (Fig. 3 C and F). In addition, the human expressed sequence tags were obtained from the brain of an infant, the cDNA used for the RACE was synthesized from the mRNA of a 37-year-old male, and the cDNA library clones were synthesized from the frontal cortex mRNA of an adult male of unspecified age or of an 85-year-old female. Therefore, BNaC1 and BNaC2 are probably expressed throughout life.
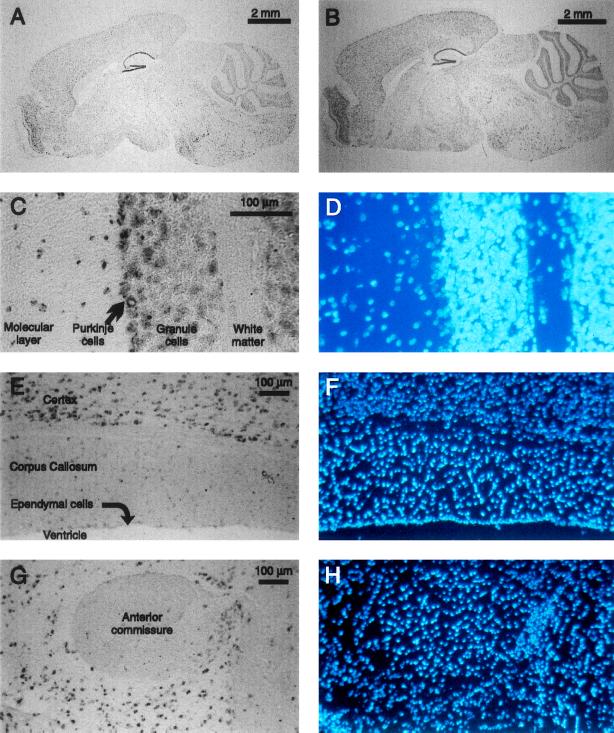
In situ hybridization in mouse sagittal brain sections (Fig. 4) revealed that all regions of gray matter coexpress both genes. The strongest expression levels are in the Purkinje and (to a lesser extent) granule cell layers of cerebellum, in dentate gyrus and regions CA1–CA4 of hippocampus, and in the olfactory bulb. In contrast, transcripts of either gene are absent or rare in regions without neuronal cell bodies: neither are detected above background levels in ependymal cells, anterior commissure, layer one of cortex, corpus callosum, or other white matter. Both genes also appear to be expressed at low levels in choroid plexus, an epithelial structure, but mBNaC2 expression is stronger than mBNaC1 (data not shown).
Figure 4.
In situ hybridization of mBNaC1 (A, E, and G) and mBNaC2 (B and C) riboprobes to mouse brain sagittal sections, and DAPI stain of nuclei in those same sections (D, F, and H). Glial cell nuclei are brighter. (A and B) Whole brain. (C and D) Portion of cerebellum with white matter, granule cell layer, Purkinje cell layer, and molecular layer indicated. (E and F) Portion of parietal cortex, corpus callosum, ependymal cell layer, and ventricular space. (G and H) Anterior commissure and surrounding neuronal areas. Hybridization of control sense riboprobes to adjacent sections under the same conditions gives no signal (not shown). In all pictures (except for C and D, whose orientation has not been determined), anterior is to the left and ventral is down.
Partial cDNA clones corresponding largely to 3′ UTRs (see Materials and Methods), which diverge between these genes, were used for fluorescent in situ hybridization on human chromosomes. hBNaC1 was mapped to 17q11.2-12 and hBNaC2 to 12q12 (Fig. 5).
Figure 5.
Mapping of hBNaC1 (A–C) and hBNaC2 (D–F) genes. FISH of cDNA fragments to human chromosomes (A and D), DAPI staining of the same chromosomes (B and E), and distribution of labeled sites in idiograms (C and F). Each dot represents double fluorescent in situ hybridization signals detected on the chromosome.
DISCUSSION
Phylogeny and Nomenclature.
The sequences of BNaC1 and BNaC2 indicate that they are clearly members of the DEG/ENaC superfamily. They are not, however, members of the degenerin branch of the superfamily: they are missing certain conserved sequences characteristic of that branch, and a phylogenetic analysis places them by themselves in a new branch of the family (refs. 14 and 23; Fig. 1). The degenerin branch thus far remains restricted to nematodes.
BNaC1 has been previously identified using the same cloning strategy (12, 13). Price et al. (12) named the protein BNC1 (for brain Na+ channel), which we prefer to write as BNaC1 both for similarity to the other superfamily members, ENaC and FaNaC, and to indicate sodium permeability. Waldmann et al. (13) found that introduction of certain mutations (modeled after the degeneration causing mutations of the nematode degenerins) in this protein could cause the death of the cells that express it, so they named it MDEG (for mammalian degenerin). Because the normal function of the protein is most likely not to cause degeneration of the cells, because mutations in certain other channels that are not degenerins can also cause degeneration (37–39), and because it is not of the degenerin branch of the superfamily, BNaC seems more functionally descriptive.
BNaC2 is novel. This protein is 68% identical to BNaC1 in amino acid sequence over its entire length, which represents greater similarity than any other two members of the DEG/ENaC superfamily (14). Several BNaC2 clones contained an insert that would encode 46 additional amino acids, situated toward the beginning of the second predicted transmembrane domain. This alternative splice exon fragment might alter the function of the protein considerably: it is in or near the pore region and immediately adjacent to a sterically constrained glycine, mutation of which can cause a degeneration phenotype (13, 21). Although such an alternative exon has not been found in any other member of the DEG/ENaC superfamily, others have been described in deg-1 (23) and α-ENaC (40). The alternate α-ENaC messages translate into proteins lacking the second transmembrane domain and do not form functional channels. They probably do not contribute to the pore, but may be regulatory subunits of a multimeric channel.
Expression.
Both BNaC1 and BNaC2 are expressed primarily in brain but not in the other organs tested. Price et al. (12) and Waldmann et al. (13) found similar results with BNaC1 by Northern blot analysis. In situ hybridization showed that, within the brain, BNaC1 and BNaC2 are expressed primarily in neurons, but are either absent from white matter or present in low amounts. An exception is the expression of BNaC2 in choroid plexus. Northern blots indicated that the expressions of BNaC1 and BNaC2 are similar but not identical among human brain regions, and that BNaC1 appears slightly earlier in mouse development than BNaC2. Hippocampal, cerebellar, and cortical regions express larger amounts of BNaC1 and BNaC2 than subcortical structures. Some neurons, such as cerebellar Purkinje cells, had higher levels of expression than others such as granule cells. On the whole, however, the expression is rather uniform among most neuronal populations.
Subunits of an Ion Channel.
It seems likely that BNaC1 and BNaC2 are ion channel subunits. Other members of the superfamily, the ENaCs and FaNaC, form sodium-permeable ion channels when expressed in frog oocytes (5–9, 11), and domain-swap experiments suggest that the degenerin MEC-4 also forms part of a channel (10, 41). Indeed, BNaC1 can form a channel by itself: injection of BNaC1 cDNA directly into oocyte nuclei (to increase expression) resulted in a small amiloride-blockable membrane current (12). In other experiments, sodium-selective currents were observed in cells that expressed a mutated BNaC1 protein (13). The mutations were equivalent to those in the degenerins that cause cell swelling; they substitute a small amino acid for a larger one in the second hydrophobic region (21, 23, 42). These mutations apparently reduce the sodium selectivity of the homomeric BNaC1 channel, from >10:1 (Na+:K+; ref. 12) to about 4:1 (13), indicating that this residue influences the pore.
On the other hand, currents are not easily elicited with BNaC1 alone: Xenopus oocytes injected with wild-type rat BNaC1 cRNA and HEK cells transfected with wild-type rat or human BNaC1 cDNA did not express detectable currents (ref. 13; unpublished results). Price et al. (12) observed small currents after nuclear injection of hBNaC1 cDNA into oocytes, which may be a more efficient method of expressing this channel, but their clone, obtained by PCR, differs in codon 495 from the various clones reported here and elsewhere (ref. 13; Fig. 2). Perhaps the Thr-495-Ala mutation, like the degeneration-causing mutations, activates a channel that would otherwise be closed. It may be that the wild-type channel is rarely open without a stimulus that is lacking in these expression systems. It may also be that BNaC1 and BNaC2 are subunits of a heteromultimeric ion channel, which requires both subunits and perhaps others to form a full-conductance channel. This would be similar to the situation with the ENaCs, in which αENaC expressed alone causes small currents, but allows much larger currents if coexpressed with β and γ ENaC (7). Similarly, genetic interaction experiments suggest that some of the degenerins form heteromultimeric channels, and that these channels contain more than one subunit of each type (22, 23, 41, 42). The variability in expression of BNaC1 and BNaC2 among brain regions may indicate differences in stoichiometry of subunits within the complete channel.
Function.
The function of the BNaC family of ion channels remains unknown. Phylogenetically, the BNaCs are equally divergent from the other branches of the DEG/ENaC superfamily, which includes channels with different functions (Fig. 1). Thus, the BNaCs are as likely to serve the same role as any other superfamily member, or a novel role. The ENaCs form a constitutively active sodium channel in kidney epithelia. Although BNaC2 is expressed in choroid plexus, an epithelium that functionally and structurally resembles that of the kidney, BNaC1 and BNaC2 are primarily expressed in neurons. Many neurons have a constant leak of sodium that contributes to their resting potential; the BNaCs may elicit this small but continuous sodium influx. As proposed for the degenerins, the BNaCs might instead form mechanically gated ion channels; such channels may regulate cell volume, a pressing matter for cells within the cranium. Or perhaps, since the FaNaC channel found in snails is activated by the FMRFamide peptide, the BNaCs may form peptide-receptor channels for one of the many neuropeptides present in the mammalian brain. Whatever their role, the distribution of BNaCs throughout the brain is striking and suggests a global function that many or all neurons share.
Acknowledgments
We thank Emily R. Liman for helpful discussions and Oksana Berezovskaja for providing brain sections. This work was supported by the Ramón Areces Foundation (to J.G.-A.), by the National Institutes of Health (NIA AG08487 to B.T.H.), and by the Howard Hughes Medical Institute (to D.P.C.). D.P.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations: UTR, untranslated region; CDS, coding sequence. Data deposition: The sequences reported in this paper have been deposited in the GenBank data base [accession nos. U57352U57352 (hBNaC1), U57353U57353 (mBNaC1), U78179U78179 (mBNaC2), and U78180U78180 or U78181U78181 (hBNaC2, with or without one alternatively spliced exon fragment)].
References
- 1.Canessa C M, Merillat A M, Rossier B C. Am J Physiol. 1994;267:C1682–C1690. doi: 10.1152/ajpcell.1994.267.6.C1682. [DOI] [PubMed] [Google Scholar]
- 2.Renard S, Lingueglia E, Voilley N, Lazdunski M, Barbry P. J Biol Chem. 1994;269:12981–12986. [PubMed] [Google Scholar]
- 3.Snyder P M, McDonald F J, Stokes J B, Welsh M J. J Biol Chem. 1994;269:24379–24383. [PubMed] [Google Scholar]
- 4.Lai C-C, Hong K, Chalfie M, Driscoll M. J Cell Biol. 1996;133:1071–1081. doi: 10.1083/jcb.133.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canessa C M, Horisberger J-D, Rossier B C. Nature (London) 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 6.Lingueglia E, Voilley N, Waldmann R, Lazdunski M, Barbry P. FEBS Lett. 1993;318:95–99. doi: 10.1016/0014-5793(93)81336-x. [DOI] [PubMed] [Google Scholar]
- 7.Canessa C M, Schild L, Buell G, Thorens B, Gautschl I, Horisberger J-D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 8.Voilley N, Lingueglia E, Champigny G, Mattei M-G, Waldmann R, Lazdunski M, Barbry P. Proc Natl Acad Sci USA. 1994;91:247–251. doi: 10.1073/pnas.91.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Nature (London) 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 10.Waldmann R, Champigny G, Lazdunski M. J Biol Chem. 1995;270:11735–11737. doi: 10.1074/jbc.270.20.11735. [DOI] [PubMed] [Google Scholar]
- 11.Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M. J Biol Chem. 1995;270:27411–27414. doi: 10.1074/jbc.270.46.27411. [DOI] [PubMed] [Google Scholar]
- 12.Price M, Snyder P, Welsh M J. J Biol Chem. 1996;271:7879–7882. doi: 10.1074/jbc.271.14.7879. [DOI] [PubMed] [Google Scholar]
- 13.Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. J Biol Chem. 1996;271:10433–10436. doi: 10.1074/jbc.271.18.10433. [DOI] [PubMed] [Google Scholar]
- 14.Corey D P, García-Añoveros J. Science. 1996;273:323–324. doi: 10.1126/science.273.5273.323. [DOI] [PubMed] [Google Scholar]
- 15.Palmer L G. Annu Rev Physiol. 1992;54:51–66. doi: 10.1146/annurev.ph.54.030192.000411. [DOI] [PubMed] [Google Scholar]
- 16.Li X-J, Blackshaw S, Snyder S H. Proc Natl Acad Sci USA. 1994;91:1814–1818. doi: 10.1073/pnas.91.5.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benos D J, Saccomani G, Sariban-Sohraby S. J Biol Chem. 1987;262:10613–10618. [PubMed] [Google Scholar]
- 18.Smith P R, Saccomani G, Joe E-H, Angelides K J, Benos D J. Proc Natl Acad Sci USA. 1991;88:6971–6975. doi: 10.1073/pnas.88.16.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staub O, Verrey F, Kleyman T R, Benos D J, Rossier B C, Kraehenbuhl J-P. J Cell Biol. 1992;119:1497–1506. doi: 10.1083/jcb.119.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalfie M, Wolinsky E. Nature (London) 1990;345:410–415. doi: 10.1038/345410a0. [DOI] [PubMed] [Google Scholar]
- 21.Driscoll M, Chalfie M. Nature (London) 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 22.Huang M, Chalfie M. Nature (London) 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- 23.García-Añoveros J, Ma C, Chalfie M. Curr Biol. 1995;5:441–448. doi: 10.1016/s0960-9822(95)00085-6. [DOI] [PubMed] [Google Scholar]
- 24.Chalfie M, Sulston J. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 25.Herman R K. Genetics. 1987;116:377–388. doi: 10.1093/genetics/116.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitani S, Du H, Hall D H, Driscoll M, Chalfie M. Development (Cambridge, UK) 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 27.Ausubel F M, Brent R, Kingston R E, Moor D D, Siedman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1988. [Google Scholar]
- 28.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 30.Panoskaltsis-Mortari A, Bucy R P. Biotechniques. 1995;18:300–306. [PubMed] [Google Scholar]
- 31.Heng H H Q, Squire J, Tsui L-C. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heng H H Q, Tsui L-C. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 33.Heng H H Q, Tsui L-C. In: Methods in Molecular Biology: In Situ Hybridization Protocols. Choo K H A, editor. Clifton, NJ: Humana; 1994. pp. 35–49. [Google Scholar]
- 34.Liu J, Schrank B, Waterston R. Science. 1996;273:361–364. doi: 10.1126/science.273.5273.361. [DOI] [PubMed] [Google Scholar]
- 35.Garnier J, Osguthorpe D J, Robson B. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 36.Chou P Y, Fasman G D. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 37.Treinin M, Chalfie M. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 38.Patil N, Cox D, Bhat D, Faham M, Myers R M, Peterson A S. Nat Genet. 1995;11:126–129. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 39.Navarro B, Kennedy M E, Velimirovic B, Bhat D, Peterson A S, Clapham D E. Science. 1996;272:1950–1953. doi: 10.1126/science.272.5270.1950. [DOI] [PubMed] [Google Scholar]
- 40.Li X-J, Xu R-H, Guggino W B, Snyder S H. Mol Pharmacol. 1995;47:1133–1140. [PubMed] [Google Scholar]
- 41.Hong K, Driscoll M. Nature (London) 1994;367:470–473. doi: 10.1038/367470a0. [DOI] [PubMed] [Google Scholar]
- 42.Shreffler W, Magardino T, Shekdar K, Wolinsky E. Genetics. 1995;139:1261–1272. doi: 10.1093/genetics/139.3.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]