Abstract
The neural cell adhesion molecule (N-CAM) mediates cell–cell interactions and is expressed in characteristic spatiotemporal patterns during development. In previous studies of factors that control N-CAM gene expression, we identified a binding site for the paired domain of Pax proteins (designated PBS) in the mouse N-CAM promoter. In this study, we demonstrate that a transcription factor known to be important for development of the central nervous system, Pax-6, binds to the N-CAM PBS and show that the PBS can influence N-CAM expression in vivo. Pax-6, produced in COS-1 cells, bound to the PBS through two half-sites, PBS-1 and PBS-2; mutations in both of these sites completely disrupted binding. Moreover, nuclear extracts from embryonic day (E) 11.5 mouse embryos bound to the PBS, and this binding was inhibited by antibodies to Pax-6. To determine the role of the PBS in vivo, we generated transgenic mice with N-CAM promoter/lacZ gene constructs containing either a wild-type or a mutated PBS. Mutations in PBS-1 and PBS-2 decreased the extent of β-galactosidase expression in the mantle layer of the spinal cord limiting it to ventral regions at E11.5. At E14.5, these mutations eliminated most of the expression that was seen in the wild-type spinal cord. Taken together with our previous observations that the PBS binds multiple Pax proteins, the data indicate that such binding contributes to the regulation of N-CAM gene expression during neural development.
Keywords: Pax-6, gene regulation, neural development, transgenic mice
The neural cell adhesion molecule (N-CAM) is a member of the immunoglobulin superfamily that is expressed in precise patterns in the nervous system and in a variety of other tissues during development (1, 2). N-CAM is expressed in the neuroectoderm shortly after neural induction and regulation of its expression has been correlated with the migration and differentiation of neural crest cells (3, 4). Alterations in the function and expression of N-CAM result in perturbations of tissue patterns during development and of nerve regeneration in the adult (5–7). These findings suggest that identification of the factors that regulate the spatiotemporal expression of N-CAM could provide significant insights into the role of this molecule in neural morphogenesis.
We and others (8–11) have identified important cis regulatory elements that control the place-dependent expression of N-CAM during development. The promoter for the mouse N-CAM gene contains several homeodomain binding sites (HBS) that interact with a number of homeodomain transcription factors including HoxB-9, HoxB-8, HoxC-6, Phox-2, and Cux (9–11). These proteins were shown in cellular cotransfection experiments to activate or repress N-CAM gene expression. Moreover, mutations in the HBS sequences altered the expression of an N-CAM promoter/lacZ fusion gene in the spinal cord of transgenic mice (12). These data support the hypothesis that the N-CAM promoter is a target for several homeodomain proteins that control the spatiotemporal pattern of N-CAM expression during embryogenesis (13, 14).
Recently, we have examined regulation of the N-CAM gene by members of the Pax family of transcription factors (15, 16), which contain a 128-amino acid DNA binding structure called the paired domain that was first described in the paired gene of Drosophila melanogaster (17). The paired domain binds to a GTTCC motif in the promoter for the Drosophila even skipped gene (18) and to a 21-bp consensus sequence (19, 20). Four Pax proteins (Pax-3, -4, -6, and -7) also contain a homeodomain that can recognize classical HBS sequences containing ATTA motifs (21).
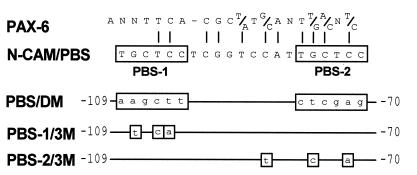
To identify DNA elements in the N-CAM promoter that bind to the paired domain of Pax proteins, we initially examined binding and regulation of the N-CAM promoter by Pax-8—a protein that contains a paired domain, but no homeodomain. A binding site for Pax-8 was identified upstream of the AUG codon between −101 and −81 in the N-CAM promoter (16). This region contained two TGCTCC motifs, designated PBS-1 and PBS-2 (for paired domain binding sites), which are similar to the GTTCC motif shown previously to bind the paired domain of Pax-1 and Pax-3 (22).
In the present study, we examine the contribution of the PBS to binding and activation of the N-CAM promoter by Pax-6, a protein containing both a paired domain and a homeodomain. To examine further the role of the PBS in vivo, we produced transgenic mice expressing the lacZ gene under the control of a wild-type N-CAM promoter or an N-CAM promoter containing mutations in the PBS. The results of these studies indicate that the PBS plays a key role in the regulation of N-CAM gene expression during neural development.
MATERIALS AND METHODS
To express Pax-6 in COS-1 cells, the Pax-6 cDNA was inserted into the SRα expression vector (23) downstream of the hemagglutinin (HA) tag. Cells were transfected with 10 μg of the Pax-6HA plasmid using lipofectamine (GIBCO/BRL), harvested 72 h posttransfection, and lysed in 100 μl of buffer (24). Protein concentration of the extracts was determined as described (25). Nuclear extracts were prepared as described (26) from the head and spinal cord of embryonic day (E) 11.5 mouse embryos.
Probes containing the wild-type (N-CAM/PBS) or mutated (PBS/DM, PBS-1/3M, and PBS-2/3M) PBS sequences (see Fig. 1) were constructed from 40-bp double-stranded oligonucleotides. Annealed oligonucleotides (5 pmol) were radiolabeled using the Klenow fragment of Escherichia coli DNA polymerase and [32P]dCTP (3000 Ci/mmol; 1 Ci = 37 GBq) (DuPont/NEN). Probes were separated by electrophoresis on a 10% polyacrylamide gel, eluted, and resuspended in H2O at 25,000 cpm/μl. Electrophoretic mobility shift assays were performed with 25,000 cpm of probe and 5 μg of Pax-6HA-transfected or mock-transfected COS-1 cell extracts in binding buffer [10 mM Hepes/200 mM KCl/0.5 mM DTT/0.1% Nonidet P-40/100 ng/μl poly(dI·dC)/10 μg bovine serum albumin]. Components were incubated at room temperature for 30 min and separated by electrophoresis on a 5% polyacrylamide gel in 0.25× TBE at 300 V for 2.5 h. Competitors were added at either 100-, 250-, or 500-fold molar excess. For supershift assays, 1 μl of a mAb to the HA tag was added to the binding mixture. Binding reactions involving embryonic nuclear extracts were performed by incubating 10 μg of nuclear extract with 50,000 cpm of PBS probe as described above.
Figure 1.
Comparison of the PBS in the N-CAM promoter to a consensus binding sequence for Pax-6. The TGCTCC motifs comprising PBS-1 and PBS-2 are highlighted within a box. Homology between the PBS and a Pax-6 consensus sequence selected from random oligonucleotides (20) is indicated by vertical lines. Base pair substitutions in the mutant probes PBS/DM, PBS-1/3M, and PBS-2/3M are shown in boxes. PBS/DM contained mutations in both PBS-1 and PBS-2, PBS-1/3M had three mutations in PBS-1, and PBS-2/3M contained three mutations in PBS-2.
For promoter activation assays, a N2A neuroblastoma cell line permanently expressing Pax-6 was transfected with reporter gene constructs prepared in the pCATbasic plasmid (Promega). One construct contained the −414 to −15 region of the N-CAM promoter driving the chloramphenicol acetyltransferase (CAT) gene and the other contained a similar promoter with mutations in PBS-1 and PBS-2 made by site-directed mutagenesis (Bio-Rad). CMVβ, a β-galactosidase (β-gal) expression plasmid (CLONTECH), was cotransfected to provide an internal reference standard for transfection efficiency. CAT activity assays and quantitation were performed as described (16).
For transgenic mice, two N-CAM promoter/lacZ gene cassettes were constructed in the pLacF vector. Construction of the wild-type N-CAM gene construct (N-CAM/PBS+) has been described (12). The N-CAM/PBS− gene construct was prepared by site-directed mutagenesis as described above. These transgenes were introduced into the RC6 mouse genome by standard oocyte microinjection techniques (27). F0 progeny were screened for the presence of the transgene by Southern analysis and PCR as described (12). Animals positive for the transgene were mated with C57BL6 mice to establish individual lines. Embryos were collected at E11.5 and E14.5, fixed in 1% formaldehyde/0.25% glutaraldyhyde in phosphate-buffered saline for 30 min at 4°C, and washed with 0.02% deoxycholate in phosphate-buffered saline. E11.5 embryos were stained in whole-mount while E14.5 embryos were either sectioned on a cryostat or vibratome. β-Gal activity was detected as described (12). For in situ hybridization, mouse embryos were fresh frozen at −70°C. Cryosections (20 μm) were hybridized to N-CAM RNA probes as described (27). Antisense and sense digoxigenin-labeled N-CAM cRNA probes were generated by in vitro transcription of the 183–505 region of mouse N-CAM cDNA. Hybridized probes were detected with antidigoxigenin IgG Fab fragments conjugated to alkaline phosphatase and visualized with BM-purple substrate (Boehringer Mannheim).
RESULTS
Pax-6 Binds to the PBS and Activates the N-CAM Promoter.
Previously, we have shown that the N-CAM promoter is activated by the Pax-8 protein via a PBS containing two TGCTCC motifs, designated PBS-1 and PBS-2 (16). Comparison of the N-CAM PBS with the consensus binding sequence for Pax-6 (Fig. 1) reveals an identity of 68% for the entire 21 bp; this identity increases to 81% when the comparison is restricted to PBS-1 and PBS-2.
To examine Pax-6 binding to the PBS, we tested four double-stranded probes (Fig. 1) for their ability to bind a HA-tagged Pax-6 protein (Pax-6HA) produced in COS-1 cells. The probe designated N-CAM/PBS containing the wild-type sequences of PBS-1 and PBS-2 was derived from the −109 to −70 region of the N-CAM promoter. Three variants of the PBS (PBS/DM, PBS-1/3M, and PBS-2/3M) were also prepared to examine the relative contribution of PBS-1 and PBS-2 to Pax-6 binding. PBS/DM contained mutations in both PBS-1 and PBS-2 that eliminated the TGCTCC motifs. PBS-1/3M had three mutations in PBS-1, but left PBS-2 intact. PBS-2/3M contained mutations that eliminated the TGCTCC motif of PBS-2, but left PBS-1 intact.
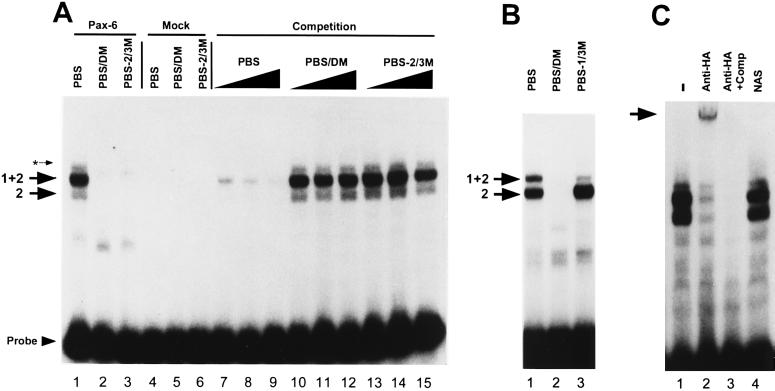
In binding assays (Fig. 2), the 32P-labeled PBS probe formed two complexes with Pax-6HA that were resolved as two bands on a polyacrylamide gel (Fig. 2A, lane 1, large arrows). Formation of both complexes was competed by addition of 100-, 250-, and 500-fold molar excess of unlabeled PBS (Fig. 2A, lanes 7, 8, and 9, respectively). 32P-labeled probes containing the PBS/DM mutation and PBS-2/3M mutation, which have an unmutated PBS-1 sequence, failed to bind Pax-6HA (Fig. 2A, lanes 2 and 3). Thus, PBS-1 alone cannot bind Pax-6. Furthermore, as competitors, PBS/DM and PBS-2/3M did not disrupt Pax-6HA binding to the PBS probe (Fig. 2A, lanes 10–15). Mutation of either the TGCTCC motif in PBS-2 or both motifs in PBS-1 and PBS-2 were therefore sufficient to eliminate Pax-6HA binding. PBS-1/3M, containing an unmutated PBS-2, bound to Pax-6HA (Fig. 2B), and 95% of the radioactivity was found in the lower band (Fig. 2B, lane 3); this indicates that PBS-1 is required for formation of the upper band. These data suggest that Pax-6 binding to PBS-2 forms the complex represented by the lower band (Fig. 2B, arrow designated 2) and binding to PBS-1 and PBS-2 form the complex represented by the upper band (Fig. 2B, arrow designated 1+2).
Figure 2.
Binding of Pax-6HA protein to the N-CAM PBS. (A) Pax-6HA expressed in COS-1 cells (lanes 1–3) and mock-transfected COS-1 cell extracts (lanes 4–6) were incubated with either 32P-labeled PBS (lanes 1, 4, and 7–15), PBS/DM (lanes 2 and 5), or PBS-2/3M (lanes 3 and 6). Complexes formed by Pax-6HA and the PBS probe are indicated by arrows labeled 2 and 1+2. A minor complex formed by both the Pax-6HA and mock-transfected COS-1 extracts is indicated by an asterisk. A 100-, 250-, or 500-fold molar excess of either unlabeled PBS (lanes 7–9), PBS/DM (lanes 10–12), or PBS-2/3M (lanes 13–15) were added as competitors. (B) Pax-6HA extracts were incubated with the PBS probe (lane 1), PBS/DM (lane 2), or PBS-1/3M (lanes 3). DNA/protein complexes are indicated by arrows labeled 2 and 1+2. (C) Supershift analysis. Pax-6HA was incubated with the 32P-labeled PBS probe and either 1 μl of HA tag antibody (lanes 2 and 3), an antibody to Ng-CAM (NAS) (lane 4), or no antibody (lane 1). In lane 3, 500-fold excess of unlabeled PBS was added as competitor. The supershifted Pax-6/PBS complex is indicated by the arrow.
To confirm that the complexes formed with the N-CAM/PBS contained the HA-tagged Pax-6 protein, supershift assays were performed using a mAb to the HA tag (Fig. 2C). Addition of the antibody resulted in the formation of a supershifted complex (Fig. 2C, lane 2) while the intensity of the bands corresponding to the Pax-6HA complexes was reduced. A mAb to an unrelated molecule (Ng-CAM) did not supershift the complexes (Fig. 2C, lane 4). Formation of the supershifted complex was competed by 500-fold molar excess of unlabeled PBS (Fig. 2C, lane 3).
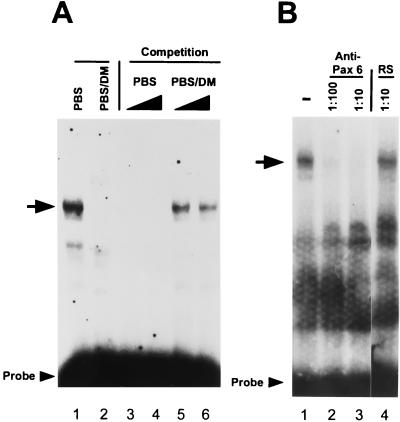
The N-CAM PBS also bound to proteins in nuclear extracts from E11.5 embryos (Fig. 3A, lane 1, arrow). This binding was inhibited by addition of an excess of unlabeled PBS (Fig. 3A, lanes 3 and 4). The PBS double mutant, PBS/DM, neither inhibited formation of this complex (Fig. 3A, lanes 5 and 6) nor bound directly to these extracts (Fig. 3A, lane 2). To determine whether Pax-6 was a component of the complex formed by embryonic nuclear extracts, we prepared a Pax-6 polyclonal antiserum and tested its ability to block or supershift the DNA/protein complex. In immunoblot analyses, this antiserum identified Pax-6 protein produced in Cos-1 cells, cross-reacted only slightly with Pax-3 and Pax-8 (1/20th of the reactivity as compared with Pax-6) (data not shown). As shown in Fig. 3B, the Pax-6 antiserum completely inhibited formation of the DNA/protein complex when either a 1:100 or 1:10 dilution was added to the binding reaction (Fig. 3B, lanes 2 and 3). Formation of this complex was not inhibited by a similar concentration of preimmune rabbit serum (Fig. 3B, lane 4). These data indicate that Pax-6 is a major component of the complex formed between the PBS and E11.5 embryonic nuclear extracts.
Figure 3.
Pax-6 in mouse embryonic nuclear extracts binds to the PBS. (A) Nuclear extracts from E11.5 embryos incubated with the PBS probe (lanes 1 and 3–6) or PBS/DM (lane 2) formed complexes indicated by an arrow. In lanes 3–6, a 100- or 250-fold molar excess of unlabeled PBS (lanes 3 and 4) or PBS/DM (lanes 5 and 6) was added as competitor. (B) Embryonic extracts were incubated with PBS probe and 5 μl of either a 1:10 (lane 2) or 1:100 (lane 3) dilution of anti-Pax-6 rabbit serum or a 1:10 dilution of normal rabbit serum (RS) (lane 4).
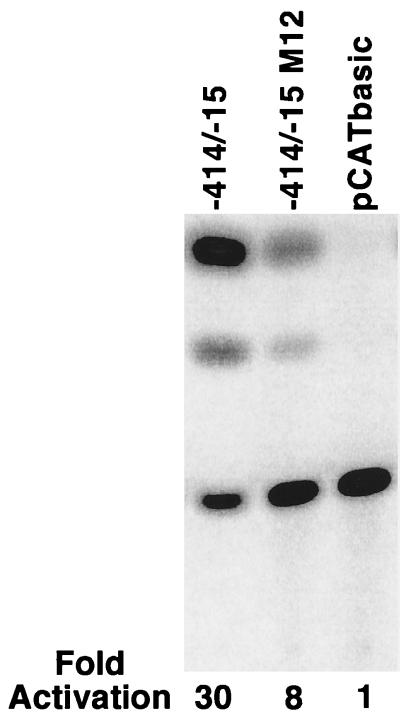
To examine the role of PBS-1 and PBS-2 in the control of N-CAM promoter activity by Pax-6, we tested two N-CAM gene constructs containing the CAT reporter gene driven either by the wild-type N-CAM promoter (−414/−15) or a similar promoter containing mutations in PBS-1 and PBS-2 (−414/−15 M12) in a N2A neuroblastoma cell line that stably expresses the Pax-6 cDNA (Fig. 4). These cells expressed 10-fold more Pax-6 mRNA than untransfected N2A cells as determined by an RNase protection assay (data not shown). As illustrated in Fig. 4, 73% of CAT activity driven by the N-CAM promoter was abolished when PBS-1 and PBS-2 were mutated. These data indicate that PBS-1 and PBS-2 are required for high levels of N-CAM promoter activity in cells expressing Pax-6.
Figure 4.
Mutations in PBS-1 and PBS-2 decrease N-CAM promoter activity in a N2A cell line stably expressing Pax-6. Cells were transfected with 0.5 μg of CMVβ and 2 μg of either a CAT gene construct driven by the −414/−15 N-CAM, a similar construct (designated −414/−15 M12) with mutations in PBS-1 and PBS-2, or the promoterless CAT gene (pCATbasic). CAT assays were performed in duplicate. The numbers refer to the relative levels of CAT activity produced by each construct.
The PBS Controls Expression of the N-CAM Gene in Vivo.
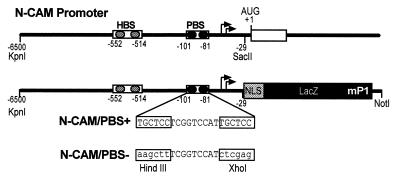
To examine the role of the PBS in place-dependent expression of N-CAM in vivo, we introduced two constructs into transgenic mice (Fig. 5). The first construct, designated N-CAM/PBS+, contained 6.5 kb of 5′ flanking sequence of the mouse N-CAM gene comprising the native PBS-1 and PBS-2, a portion of the first exon, and the bacterial lacZ gene. The second construct, designated N-CAM/PBS−, was identical to N-CAM/PBS+ except that PBS-1 and PBS-2 were replaced by the PBS/DM mutation. Three lines of N-CAM/PBS+ mice and four lines of N-CAM/PBS− mice were generated and examined for β-gal expression both in whole-mount and transverse sections. In toto, 41 N-CAM/PBS+ and 38 N-CAM/PBS− embryos were examined. In all embryos analyzed, β-gal was expressed in a subset of the tissues known to express N-CAM and was never observed in tissues outside of those that normally express N-CAM. In the head, staining was consistently observed in the developing eyes (including the lens and neural retina), the olfactory epithelia, and the mantle layer of the hindbrain and diencephalon, although the pattern and intensity varied within these structures. Some lines also showed staining in the trigeminal ganglia.
Figure 5.
Schematic representation of the 5′ flanking region of the mouse N-CAM gene showing N-CAM/PBS+ and N-CAM/PBS− transgenes. Arrows represent transcription initiation sites. The site of translation initiation is designated as +1. The sequence of the PBS, and the base pair substitutions made in the PBS-1 and PBS-2 in N-CAM/PBS− are indicated. The location of the HBS in the N-CAM promoter is also shown.
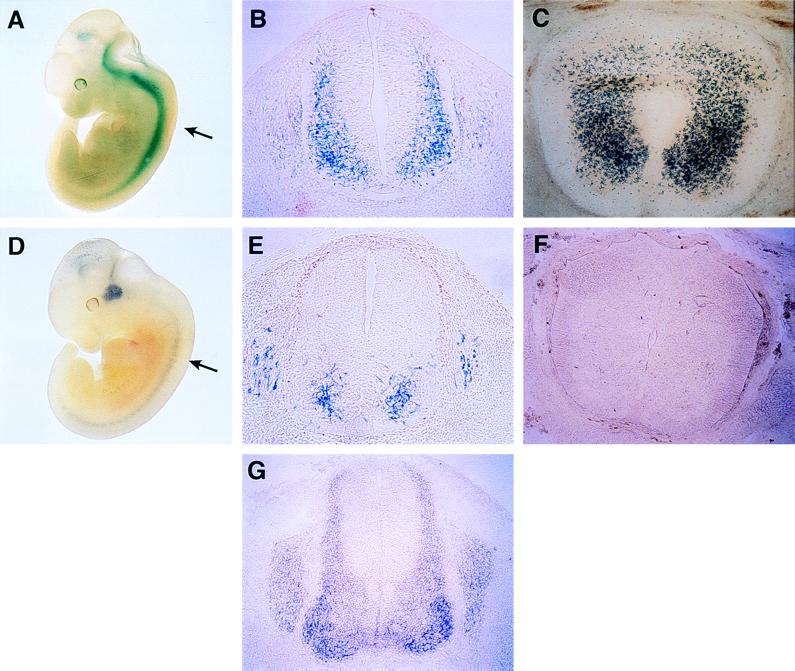
At E11.5, the overall pattern of β-gal expression in N-CAM/PBS− embryos was similar to that observed in the N-CAM/PBS+ mice in rostral regions (Fig. 6). An examination of transverse sections revealed no differences between E11.5 N-CAM/PBS+ and N-CAM/PBS− embryos in the β-gal staining pattern in the brain, eyes, and olfactory epithelia (data not shown). In the spinal cord, however, the β-gal expression pattern in N-CAM/PBS− mice was considerably different from that observed in N-CAM/PBS+ embryos. N-CAM/PBS+ embryos showed β-gal staining along the entire anteroposterior length of the spinal cord at E11.5 (Fig. 6A). In transverse sections (Fig. 6B) β-gal staining was observed throughout the mantle layer and in dorsal root ganglia. This pattern was comparable to N-CAM mRNA expression in the spinal cord as detected by in situ hybridization (Fig. 6G). In contrast, β-gal expression in E11.5 N-CAM/PBS− embryos (Fig. 6D) was limited to a small population of cells in the most ventral region of the spinal cord (Fig. 6E). This pattern of expression did not extend into the alar plate as was observed in N-CAM/PBS+ embryos (compare Fig. 6 B with E).
Figure 6.
Expression of β-gal in N-CAM/PBS+ and N-CAM/PBS− transgenic embryos. Stained whole-mounts of E11.5 N-CAM/PBS+ (A) and N-CAM/PBS− embryos (D). Cross sections of either N-CAM/PBS+ (B and C) or N-CAM/PBS− (E and F) embryos were taken at a lumbar position indicated by the arrows in A and D at either E11.5 (B and E) or E14.5 (C and F). In situ hybridization for N-CAM mRNA at the same lumbar cross section of an E11.5 embryo is shown in G.
At E14.5, the N-CAM/PBS+ transgene was expressed in postmitotic neurons throughout the entire spinal cord (Fig. 6C). However, in N-CAM/PBS− embryos, only a few cells in the ventral portion of the spinal cord showed expression (Fig. 6F). The combined data indicate that the PBS in the N-CAM promoter is required to direct and maintain the dorsoventral pattern of N-CAM expression in the developing spinal cord.
DISCUSSION
As part of an ongoing effort to understand how the expression of CAMs is controlled during embryogenesis, we have isolated the promoters for CAM genes, identified specific cis-regulatory elements, and have begun to characterize the trans-factors that regulate such expression (9, 16, 28, 29). In the course of these studies, we identified an element in the N-CAM promoter called the PBS (16) that binds the paired domain—a DNA binding structure common to the Pax family of transcription factors that regulate neural development (22, 30, 31). The N-CAM PBS is composed of two half-sites (PBS-1 and PBS-2) and is related to the consensus binding sequence for several different Pax proteins (19, 20, 32). The PBS was originally identified (16) as a sequence that is bound by Pax-8 (a protein containing the paired domain but no homeodomain) that is known to be expressed in a rostral-caudal pattern in the metencephalon and spinal cord (22).
Here we show that the PBS also binds to Pax-6, a protein containing both a paired domain and a homeodomain that is known to be important for the development of the central nervous system, eye, and olfactory system. In transgenic mice, we found that mutations in the PBS altered expression of the N-CAM promoter in the spinal cord, indicating that a binding site for Pax proteins can influence N-CAM expression in vivo. Together with our previous work (16) the results support the conclusion that several members of the Pax gene family may regulate N-CAM expression via the PBS.
A model for Pax-6 binding to DNA has been proposed (32) which suggests that the paired domain binds as a monomer to two half-sites within the Pax-6 binding sequence. In this model, the amino-terminal portion of the paired domain makes initial contact with one half-site, resulting in a conformational change in the protein that allows interaction of the carboxyl-terminal subdomain with the other half-site. We believe that the TGCTCC motifs in PBS-1 and PBS-2 represent such half-sites. In our experiments, Pax-6 formed two prominent complexes with the PBS (upper and lower bands, Fig. 2). The TGCTCC motif of PBS-2 was required to observe any binding of Pax-6 to the PBS, indicating that PBS-2 may represent an initial contact point for Pax-6. When PBS-1 was mutated leaving only PBS-2 intact, only one complex (the lower band) was observed. Although Pax-6 could bind to PBS-2 alone, both PBS-1 and PBS-2 together were required to form the upper band. Thus, in accord with the monomer model (32), the lower complex most likely represents Pax-6 binding to PBS-2 and the upper complex represents Pax-6 binding to PBS-1 and PBS-2.
In transgenic mice, the PBS was essential for normal patterning of N-CAM gene expression in the spinal cord. At E11.5, mutations in the PBS disrupted the expression of N-CAM in the mantle layer. At E14.5, in marked contrast to the intense expression observed in lines with the wild-type N-CAM promoter, β-gal expression by the PBS mutant was barely detectable in the spinal cord. These observations demonstrate that the PBS is not only required for proper dorsoventral patterning but also for maintenance of N-CAM expression in the spinal cord. A more extensive analyses of the β-gal-expressing cells using defined neuronal and glial markers will be required to determine which cell populations lose N-CAM expression when the PBS is mutated.
All transgenic lines carrying the native N-CAM promoter showed β-gal expression in the nervous system. However, each line represented only a particular subset of the overall N-CAM expression pattern. These findings suggest that additional regulatory elements located elsewhere may be required to insulate the promoter and stabilize the neural pattern. Similar variability was observed in the N-CAM/PBS− mice. Despite this variability, the loss of N-CAM expression in the spinal cord produced by the PBS mutation was consistently found in all cases.
Pax-8 (16), Pax-6, and other Pax proteins (15) bind to and regulate the N-CAM promoter in vitro, raising the possibility that several Pax proteins may regulate N-CAM expression in vivo. Inputs from several Pax proteins may need to be perturbed to produce alterations in the pattern of N-CAM expression. Consistent with this notion, mutations in the PBS disrupted N-CAM expression in the spinal cord where multiple Pax genes are expressed, but not in the rostral central nervous system where only Pax-6 is expressed (33–35). Recent binding studies in our laboratory also indicate that at least one other site upstream of the PBS binds to the paired domain of Pax-6 in a sequence-specific manner (unpublished results). Thus, mutations in combinations of Pax-6 binding sites may be required to disrupt N-CAM expression in anterior structures where Pax-6 has been observed to have phenotypic effects (36–38). It has been found, for example, that mutations in not one, but any two out of three, binding sites for the homeodomain protein Pbx-1 are necessary to eliminate expression of the HoxB1 gene in the fourth rhombomere during hindbrain development (26).
Pax-6 may also bind via its homeodomain to other sites in the N-CAM promoter. Pax proteins such as Pax-6 having paired homeodomains can form dimers on classical HBS containing the TAAT motif (21) and have been proposed to engage in multiple binding modes depending on the presence or availability of PBS and HBS sequences within target promoters (39). Although Pax-6 does not appear to bind directly to the HBS in the N-CAM promoter (unpublished results), it may interact with proteins bound to the HBS. These proteins include members of the Hox family and Phox-2 which are known to bind to the HBS and regulate N-CAM promoter activity (9–11). These various interactions may lead to unique patterns of N-CAM expression during embryogenesis. In this connection, it is pertinent that mutations in either the HBS (12) or the PBS both lead to the ventralized expression of lacZ in the spinal cord at E11.5 and cause an absence of lacZ expression at E14.5. These findings suggest that the HBS and PBS may be required either individually or combinatorially to ensure proper expression of N-CAM in the spinal cord.
While this and our previous studies suggest that the PBS in the N-CAM gene is a target of Pax proteins, and that mutations in the PBS disrupt the neural expression of N-CAM, it remains a challenge to identify the particular Pax proteins that carry out patterning in vivo. To accomplish this, it will be necessary to breed mice carrying the wild-type N-CAM promoter/lacZ mice to mice in which specific Pax genes are mutated, deleted, or overexpressed and then examine alterations of N-CAM expression in these mutant backgrounds. It should be particularly informative to mate the N-CAM/PBS+ mouse with the Small eye mouse (36) containing mutations in the Pax-6 gene.
Acknowledgments
We are grateful to Madhavi Katragadda and Brett Fenson for excellent technical assistance and Farid Karimi for assistance with animal care. We thank Drs. K. Crossin, B. Cunningham, L. Krushel, and V. Mauro for critical reading of the manuscript. This work was supported by U.S. Public Health Service Grants HD33576 to G.M.E. and NS34493 to F.S.J., and by a grant from the G. Harold and Leila Y. Mathers Charitable Foundation to G.M.E. G.M.E. is a consultant to Becton Dickinson and Company.
Footnotes
Abbreviations: N-CAM, neural cell adhesion molecule; HBS, homeodomain binding site(s); β-gal, β-galactosidase; PBS, paired domain binding site; HA, hemagglutinin; E, embryonic day; CAT, chloramphenicol acetyltransferase.
References
- 1.Crossin K L, Chuong C-M, Edelman G M. Proc Natl Acad Sci USA. 1985;82:6942–6946. doi: 10.1073/pnas.82.20.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edelman G M, Crossin K L. Annu Rev Biochem. 1991;60:155–190. doi: 10.1146/annurev.bi.60.070191.001103. [DOI] [PubMed] [Google Scholar]
- 3.Thiery J-P, Duband J-L, Rutishauser U, Edelman G M. Proc Natl Acad Sci USA. 1982;79:6737–6741. doi: 10.1073/pnas.79.21.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuong C-M, Edelman G M. J Cell Biol. 1985;101:1009–1026. doi: 10.1083/jcb.101.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser S E, Carhart M S, Murray B A, Chuong C-M, Edelman G M. Dev Biol. 1988;129:217–230. doi: 10.1016/0012-1606(88)90176-5. [DOI] [PubMed] [Google Scholar]
- 6.Rieger F, Nicolet M, Pincon-Raymond M, Levi G, Edelman G M. J Cell Biol. 1988;107:707–719. doi: 10.1083/jcb.107.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniloff J K, Levi G, Grumet M, Rieger F, Edelman G M. J Cell Biol. 1986;103:929–945. doi: 10.1083/jcb.103.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch M-R, Gaugler L, Deagostini-Bazin H, Bally-Cuif L, Goridis C. Mol Cell Biol. 1990;10:1959–1968. doi: 10.1128/mcb.10.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones F S, Prediger E A, Bittner D A, De Robertis E M, Edelman G M. Proc Natl Acad Sci USA. 1992;89:2086–2090. doi: 10.1073/pnas.89.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones F S, Holst B D, Minowa O, De Robertis E M, Edelman G M. Proc Natl Acad Sci USA. 1993;90:6557–6561. doi: 10.1073/pnas.90.14.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valarché I, Tissier-Seta J-P, Hirsch M-R, Martinez S, Goridis C, Brunet J-F. Development (Cambridge, UK) 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Jones F S, Krushel L A, Edelman G M. Proc Natl Acad Sci USA. 1996;93:1892–1896. doi: 10.1073/pnas.93.5.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelman G M. Dev Dyn. 1992;193:2–10. doi: 10.1002/aja.1001930103. [DOI] [PubMed] [Google Scholar]
- 14.Edelman G M, Jones F S. Philos Trans R Soc London B. 1995;349:305–312. doi: 10.1098/rstb.1995.0118. [DOI] [PubMed] [Google Scholar]
- 15.Chalepakis G, Jones F S, Edelman G M, Gruss P. Proc Natl Acad Sci USA. 1994;91:12745–12749. doi: 10.1073/pnas.91.26.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holst B D, Goomer R S, Wood I C, Edelman G M, Jones F S. J Biol Chem. 1994;269:22245–22252. [PubMed] [Google Scholar]
- 17.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 18.Chalepakis G, Fritsch R, Fickensher H, Deutsch V, Goulding M, Gruss P. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- 19.Czerny T, Schaffner G, Busslinger M. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 20.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 21.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 22.Chalepakis G, Stoykova A, Wijnholds J, Tremblay P, Gruss P. J Neurobiol. 1993;24:1367–1384. doi: 10.1002/neu.480241009. [DOI] [PubMed] [Google Scholar]
- 23.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 24.Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbànek P, Maurer-Fogy I, Busslinger M. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 25.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Pöpperl H, Bienz M, Studer M, Chan S-K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 27.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 28.Goomer R S, Holst B D, Wood I C, Jones F S, Edelman G M. Proc Natl Acad Sci USA. 1994;91:7985–7989. doi: 10.1073/pnas.91.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallunki P, Jenkinson S, Edelman G M, Jones F S. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 30.Mansouri A, Stoykova A, Gruss P. J Cell Sci. 1994;18:35–42. doi: 10.1242/jcs.1994.supplement_18.5. [DOI] [PubMed] [Google Scholar]
- 31.Chalepakis G, Tremblay P, Gruss P. J Cell Sci Suppl. 1992;16:61–67. doi: 10.1242/jcs.1992.supplement_16.8. [DOI] [PubMed] [Google Scholar]
- 32.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 33.Plachov D, Chowdhury K, Walther C, Simon D, Guenet J-L, Gruss P. Development (Cambridge, UK) 1990;110:643–651. doi: 10.1242/dev.110.2.643. [DOI] [PubMed] [Google Scholar]
- 34.Goulding M D, Walther C, Joste B, Plachov D, Gruss P. Expression of Developmentally Regulated Genes in the Embryonic Mouse Nervous System. Amsterdam: Elsevier; 1991. [Google Scholar]
- 35.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 36.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson J M, Prosser J, Jordan T, Hastie N D, vanHeyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 37.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, vanHeyningen V. Nat Genet. 1992;1:328–332. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 38.Ton C C T, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, vanHeyningen V, Hastie N D, Meijers-Heijboer H, Dreschsler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Saunders G F. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 39.Jun S, Desplan C. Development (Cambridge, UK) 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]