Abstract
Membrane excitability in different tissues is due, in large part, to the selective expression of distinct genes encoding the voltage-dependent sodium channel. Although the predominant sodium channels in brain, skeletal muscle, and cardiac muscle have been identified, the major sodium channel types responsible for excitability within the peripheral nervous system have remained elusive. We now describe the deduced primary structure of a sodium channel, peripheral nerve type 1 (PN1), which is expressed at high levels throughout the peripheral nervous system and is targeted to nerve terminals of cultured dorsal root ganglion neurons. Studies using cultured PC12 cells indicate that both expression and targeting of PN1 is induced by treatment of the cells with nerve growth factor. The preferential localization suggests that the PN1 sodium channel plays a specific role in nerve excitability.
Keywords: dorsal root ganglia, PC12 cells, nerve growth factor, superior cervical ganglia, neurites
Excitable cells have in common the ability to generate action potentials. In nerve and muscle cells, this ability is due to the presence of voltage-gated sodium channels in the plasma membrane that permit the rapid entry of sodium ions into the cell during depolarization (1). The voltage-dependent sodium channel in mammals consists of a macromolecular assembly of α and β subunits, with the α subunit being the pore-forming component (reviewed in ref. 2). The α subunit is encoded by a large family of related genes. Several of the α subunit mRNAs are distributed broadly within the central nervous system (CNS; brain types I, II, III, VI; refs. 3–6), whereas other types are restricted to muscle (7–9).
Previous molecular and biochemical studies have indicated that sodium channel types distinct from those in CNS and muscle exist in the peripheral nervous system (PNS). Northern blot analyses, using a probe encoding highly conserved sodium channel sequences, detect an abundant sodium channel mRNA of a size unique to the PNS (10, 11). Similarly, antibody studies indicate the presence of a sodium channel type in the PNS that is not accounted for by the other known CNS sodium channel types (12). Physiology studies indicate that multiple sodium channel types are present in the PNS. For example, at least two distinct sodium currents with different sensitivities to the blocker tetrodotoxin (TTX) have been observed in dorsal root ganglia (DRG) (13–15). Candidates for these distinct sodium channel types are beginning to emerge. Recently, a new sodium channel α subunit type has been identified, PN3/SNS, that is localized to a small population of sensory neurons in DRG, and that gives rise to TTX-insensitive sodium currents when expressed in Xenopus oocytes (16, 17). Because of these features, it is likely that the PN3/SNS sodium channel is responsible for the TTX-insensitive current observed in some sensory neurons. The relative amount of PN3/SNS mRNA in the ganglia, deduced from RNA hybridization data, suggests that this sodium channel is not a major subtype in the PNS ganglia. This finding, coupled with the observation that the predominant sodium current in adult DRG neurons is TTX sensitive (13–15), makes it likely that a major sodium channel type in PNS is still not identified.
We described previously a putative sodium channel mRNA, of a novel size, that was induced by treatment of PC12 cells with nerve growth factor (NGF) [peripheral nerve 1 sodium channel (PN1); refs. 11 and 18). Because NGF-differentiated PC12 cells are a widely used model for peripheral sympathetic neurons, the new sodium channel mRNA was a candidate for coding for the major TTX-sensitive sodium current in PNS. We have now identified the full-length cDNA coding for the PN1 α subunit and have examined in detail its cellular and subcellular distributions in peripheral neurons.
MATERIALS AND METHODS
Cell Culture.
PC12 cells (19) and the PKI-4 PC12 subclone that expresses the cAMP-dependent kinase inhibitor protein (11) were grown on tissue culture dishes in Dulbecco’s modified Eagle’s medium (GIBCO/BRL) supplemented with 10% donor horse serum (JRH Biosciences, Lenexa, KS), 5% fetal bovine serum (JRH Biosciences), and 1% penicillin/streptomycin (GIBCO/BRL), in an atmosphere of 10% CO2 at 37°C. β-NGF was purified from mouse submaxillary glands (20) and used at a final concentration of 100 ng/ml.
PCR Amplification.
Total cellular RNA was isolated according to the method of Cathala et al. (21) from NGF-treated PKI-4 cells. Two micrograms of the total RNA was used to synthesize first-strand cDNA using random hexamer primers for the reverse transcriptase reaction (22). The cDNA then served as template for the PCR amplification, using a pair of degenerate oligonucleotide primers that specified a 400-bp region within repeat domain III of the sodium channel α subunit gene (22). The PCR products were restricted, excised from a low-melt agarose gel (SeaPlaque GTG, FMC), and subcloned into a Bluescript II SK plasmid vector cleaved previously with HindIII and BamHI. The clones were screened for cDNA inserts by miniprep (23) and sequenced in both directions by dideoxy chain termination (Sequenase 2.0 kit, United States Biochemical). Sequence data were compiled and analyzed using GeneWorks software (IntelliGenetics). From this PCR reaction, a 400-bp product termed PC12–1 was obtained that encodes a portion (amino acids 1347–1477 in the brain type II sodium channel) of the PN1 sodium channel.
cDNA Library Construction and Screening.
Poly(A)+ mRNA from the PKI-4 PC12 subclone was purified (mRNA purification kit, Pharmacia) and used to construct a random and oligo(dT)-primed λ ZAPII cDNA library (Stratagene). The library consisted of ≈1.4 × 107 independent clones prior to amplification. Screening of ≈8 × 105 recombinants with a 32P-labeled cDNA probe (oligo labeling kit, Pharmacia) generated from either pPC12–1 or the 5′ ends of derived cDNAs resulted in the isolation of clones PN1–7, PN1–17, PN1–24, PN1–42, and PN1–49. Two of the clones, PN1–49 and PN1–42 (spanning nt 1–3063 and 2326–5952 of the coding region, respectively), overlap by 738 bp to generate the full-length PN1 cDNA. Plasmid PN1–49 contains 325 bp of 5′ untranslated sequence, and an additional 85 bp of 5′ untranslated sequence were found using 5′ rapid amplification of cDNA ends (data not shown). Clone PN1–42 contains 3.5 kb of 3′ untranslated sequence, including the consensus polyadenylylation signals (24), but lacks a poly(A) tail. Like other sodium channels, PN1 does not have a consensus sequence for a eukaryotic initiation site, but the 5′ untranslated region contains an out-of-frame ATG codon at position −8 to −6, which is conserved in previously cloned sodium channel cDNAs. The 5952-bp open reading frame predicts a protein with 1984 amino acids and a calculated molecular mass of 225,791 daltons (Fig. 1).
Figure 1.
Deduced primary structure of the PN1 α subunit. The putative six transmembrane helices (S1–S6) within each repeat domain (I–IV) are underlined.
Northern Blot and Ribonuclease Protection Analyses.
Total cellular RNA was isolated from PC12 cells as described in Cathala et al. (21) and from adult Sprague–Dawley rat brain, spinal cord, superior cervical ganglion, dorsal root ganglion, skeletal muscle, cardiac muscle, and adrenal gland using the standard method of Chirgwin et al. (25). Northern blot analysis was performed according to Toledo-Aral et al. (18) using 32P-UTP-labeled antisense RNA probes generated from the following linearized templates: pPC12–1, pRB211 (26), and pIB15 (cyclophilin; ref. 27). RNA probes were transcribed with either T3 (pPC12–1) or SP6 (pRB211 and pIB15) RNA polymerases. RNase protection assays were performed by use of the RPA II kit (Ambion, Austin, TX). Total RNA was hybridized with 104 cpm of antisense RNA probe generated from pPC12–1 and the internal control pTRI-beta-actin (Ambion). Autoradiography was performed at −80°C using preflashed Kodak XAR-5 film with an intensifying screen. Levels of RNA were normalized to the levels of cyclophilin mRNA (Northern blot) or β actin mRNA (RNase protection assay).
In Situ Hybridization.
Newborn and adult mice were anesthetized by hypothermia or Avertin (200 mg/kg), respectively, and perfused transcardially with 4% paraformaldehyde in PBS (pH 7.4). Embryos were anesthetized by hypothermia and immersed in the same fixative solution. All mice were postfixed in fresh fixative overnight and then cryoprotected and OCT-embedded and sectioned at 10 μm. Sections from different regions of the embryo, as well as from different regions of the brain from newborn and adult mice were selected for in situ hybridization. The in situ hybridization was performed using 33P-labeled cRNA probes generated from plasmid PC12–1 (28). Following a 1-week exposure at 4°C, slides were developed in D19 developer (Kodak), fixed in Kodak Fixer. After counterstaining with 0.5% cresyl violet, slides were dehydrated and coverslipped.
Antibodies.
Antisera were raised, by immunizing rabbits to a synthetic peptide coupled to keyhole limpet hemocyanin, to an epitope in PN1 (residues 446–460), which is not present in any of the other known sodium channels. Antibodies were affinity purified on peptide columns before use. Sodium channel antibody raised against a conserved epitope of vertebrate sodium channels, corresponding to amino acids 1473–1490 of PN1, was prepared as described (29).
Western Blots.
Whole DRG were dissected from embryonic day 15 (E15) rat embryos and homogenized using an Ultra-Turrax apparatus in a buffered protease-inhibitor solution (30). Homogenates were spun at 1500 × g for 5 min to remove nuclei, and crude membranes were removed from the supernatant after centrifuging at 100,000 × g for 1 hr. Homogenates of rat brain and muscle were prepared in the same manner. Membranes were then solubilized, subjected to SDS/PAGE, and transferred to nitrocellulose. The membranes were probed with the antibodies described above using standard procedures. As controls, membranes were probed with antibodies that were preincubated overnight with a 100-fold excess of peptide antigen.
Immunocytochemistry.
Primary cultures of E15 DRG neurons were fixed in 3.7% formaldehyde, permeabilized with methanol, and blocked in 15% goat serum in PBS, and incubated with affinity-purified PN1 specific antibodies overnight at room temperature. The cells were immunostained with donkey anti-rabbit antiserum lissamine rhodamine conjugate (The Jackson Laboratory). PC12 cells were fused using polyethyleneglycol and plated on glass coverslips coated with polylysine and laminin as described in Halegoua (31). Cells were treated with NGF 24 and 48 hr after plating. Three days after plating, PN1 was visualized by indirect immunofluorescence using the affinity-purified PN1 specific antibodies and a fluorescein-conjugated goat affinity-purified secondary antibody to rabbit IgG (Cappel), as described (31). In both cases, control cells (E15 DRG neurons and PC12) were treated with antibodies that were preincubated with a 100-fold excess of peptide antigen overnight. Confocal images were obtained using a laser scanning confocal microscope (MRC 600; Bio-Rad).
RESULTS
The PN1 Sodium Channel α Subunit Is Distinct Structurally from the Other Rat Neuronal Sodium Channel Types.
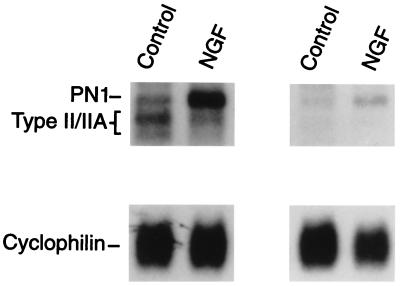
To clone the PN1 cDNA, reverse transcriptase-PCR was performed using total cellular RNA isolated from NGF-treated PKI-4 PC12 cells (11). A 400-bp cDNA (PC12–1) was isolated from the reaction products and identified as a portion of a new sodium channel by sequence analysis. To confirm the authenticity of this cDNA, we took advantage of previous findings revealing a selective induction of “PN1 mRNA” in PC12 cells treated with NGF (11, 18). In Northern blots, the putative PN1 cDNA probe hybridized specifically to the 11-kb transcript in PC12 cells and this mRNA was induced by treatment of the cells with the growth factor (Fig. 2). Using the PC12–1 cDNA as a probe for screening a library generated from NGF-treated PKI-4 PC12 cells (see Materials and Methods), two overlapping cDNA clones were isolated that together spanned the entire PN1 α subunit.
Figure 2.
An antisense RNA generated from pPC12–1 (PN1) hybridizes specifically to the 11-kb transcript in PC12 cells. Total cellular RNA (10 μg) from control and NGF-treated PC12 cells (5 hr of treatment) was subjected to Northern blot analysis. Duplicate blots were hybridized with antisense RNA probes generated from the generic sodium channel template pRB211 (Left) and from the cloned PCR product pPC12–1 (Right). The positions of the PN1, type II/IIA, and cyclophilin mRNAs are indicated.
The primary structure of the PN1 α subunit was deduced from the single open reading frame and the translational initiation site was assigned to the first in-frame ATG codon (Fig. 1). Using the presumed initiation site, the deduced PN1 primary structure can be aligned precisely with the primary structures of the other known neuronal subunits. The deduced primary structure of the PN1 α subunit is 76% identical to the brain type II α subunit and contains all of the structural hallmarks characteristic of the neuronal sodium channels. In particular, the putative pore-lining segments (SS1–SS2), which contain residues shown to be involved in sodium-selective permeation and TTX sensitivity, are present in PN1. On the other hand, PN1 exhibits some features that distinguish it from the Type I, II, and III channels. For example, the PN1 protein has fewer potential asparagine-linked (N-linked) glycosylation sites. Significant differences among PN1 and the Type I, II, and III sodium channels are found in some of the putative extracellular loops between S5 and SS1, the intracellular loop between domain I and II, and the carboxyl terminus. The functional significance of these differences remain to be clarified by further studies.
The PN1 Gene Is Expressed to High Levels Throughout the Peripheral Nervous System.
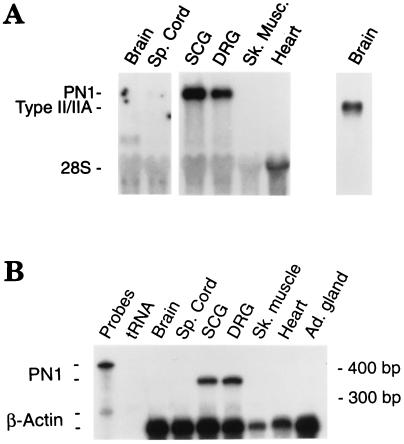
The tissue distribution of the PN1 sodium channel was first determined using RNA hybridization analyses. Northern blots using a PN1-specific antisense RNA probe (PC12–1) detected an ≈11-kb transcript that was abundant in both sensory and sympathetic ganglia. The PN1 mRNA was detected in superior cervical and DRG ganglia (Fig. 3A), and trigeminal ganglion (not shown). In contrast to these tissues, the levels of PN1 mRNA were barely detectable in spinal cord, and no transcripts were detected in skeletal muscle and cardiac muscle, brain (Fig. 3A), and liver (data not shown). RNase protection analyses confirmed the high levels of expression of PN1 in peripheral nerve tissues (Fig. 3B). Very low amounts of PN1 transcript could be detected in rat adrenal gland RNA with long exposure times (data not shown).
Figure 3.
The PN1 gene is expressed preferentially in the PNS. (A) Northern blot analysis using an antisense RNA probe from PN1 sequences (left) or from the generic sodium channel template pRB211 (far right) indicating the positions of the type II/IIA transcripts. Each lane contains 8 μg of total RNA. (B) RNase protection assay indicating that the mRNA detected in Northern blot analysis is identical to PN1. Total cellular RNA (5 μg) from the indicated tissues was hybridized to the PN1-specific antisense RNA probe (pPC12–1; 424 nt). The protected RNA fragments are the expected 343 nt. To control for differences in the amounts of RNA among samples a probe for β actin (protected fragment = 250 bp) is also included.
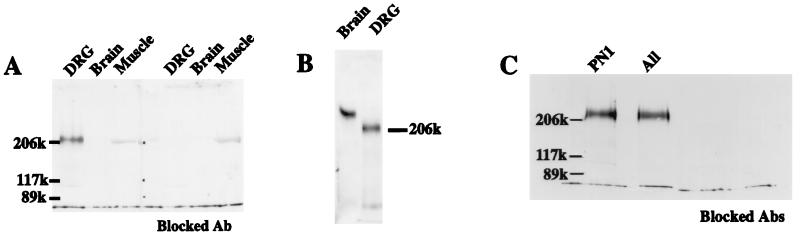
To determine that the restriction of PN1 mRNA to the PNS was also reflected in the distribution of PN1 protein, a polyclonal antibody was raised against a PN1 peptide that is not shared with other known sodium channels (see Materials and Methods). A single band of strong immunoreactivity, blocked in the presence of the immunizing peptide, was observed in Western blots of extracts of whole E15 DRG (Fig. 4A). Extracts of rat brain and skeletal muscle, which contain other sodium channel types, showed no detectable PN1 immunoreactivity at equivalent levels of applied protein. Challenge of DRG blots with a sodium channel antibody directed against an epitope conserved among different sodium channels (29) revealed only a single band of the same electrophoretic mobility as detected with the PN1-specific antibody (Fig. 4C). Thus, the protein and RNA analyses taken together indicate that PN1 is a major sodium channel type in the PNS.
Figure 4.
PN1 is a major sodium channel type in DRG. (A) Western blot analysis using membrane extract (4 μg per lane) isolated from different tissues, visualized using a PN1-specific antibody. For controls, the antibody was preincubated with the immunizing peptide (Blocked Ab). In the muscle extracts, the weak nonspecific signal, presumably myosin, is not blocked by the antibody incubation with PN1 peptide. (B) Western blot analysis using rat brain membrane extract (0.5 μg per lane) and DRG membrane extract (5 μg/lane), visualized using an antibody directed against an epitope conserved among neuronal sodium channels. (C) Western blot analysis using DRG membrane extract (4 μg per lane). The antibodies raised against the conserved sodium channel epitope (“All”) and the PN1-specific epitope (PN1) both recognize a protein of the same size. Control lanes (Blocked Abs) are as described for A.
Interestingly, the PN1 band migrated as a significantly smaller species (Mr of about 210 kDa) than the bulk of sodium channels in either rat brain (Fig. 4B) or rat muscle (not shown), as visualized using the “conserved” antibody. However, the overall length of the deduced amino acid sequence is not significantly different from either of the major sodium channel isoforms of rat brain or skeletal muscle. It is possible that the difference observed in the mobility of SDS/PAGE is related to the unusually small number of consensus sites for N-linked glycosylation in the PN1 sequence.
The PN1 α Subunit Is Localized to Neurons and Targeted to Neurite Terminals.
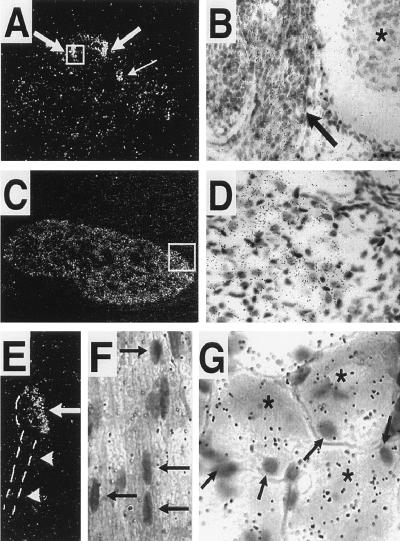
An in situ hybridization study (Fig. 5) indicated that PN1 expression was confined to the neuronal population within the peripheral ganglia. Both sensory and sympathetic ganglia contained PN1 transcripts. No differences in the patterns of expression of PN1 mRNA were observed between neonate and adult ganglia (Fig. 5 and data not shown) and, in concurrence with the RNase protection analyses, signals were never detected in brain sections (data not shown). PN1 transcripts were not detected in Schwann cells that populate the nerve roots (Fig. 5 E and F) or in satellite cells of nerve in DRG (Fig. 5G). Thus, the in situ hybridization study showed both the neuronal specificity of PN1 gene expression in the PNS and its absence in postmitotic neurons in the CNS.
Figure 5.
PN1 expression is restricted to neuronal populations within peripheral ganglia. (A) Dark-field photograph from a transverse section through an E13.5 embryo showing PN1 transcripts in DRGs (thick arrows) and sympathetic chain (thin arrow). (B) Bright-field view from the inset in A showing the DRG (arrow) and spinal cord (asterisk). (C) Coronal section through the head of a newborn mouse showing PN1 expression in the trigeminal ganglia. (D) Bright-field view from the inset in C showing PN1 transcripts with respect to the neuronal and glial cell areas within the trigeminal ganglia. (E) Dark-field photograph of an adult mouse DRG showing high levels of PN1 transcripts (arrow). Note lack of PN1 transcripts in nerve root (arrowheads). (F) Higher magnification (bright field) of an adult DRG nerve root cut longitudinally. Note lack of PN1 transcripts in the Schwann cells (arrows). (G) Higher magnification (bright field) of an adult DRG showing PN1 expression in sensory neurons (asterisks) but not in nonneuronal cells (arrows). (A, C, and E, ×250; B and D, ×450; F and G, ×800.)
The subcellular localization of PN1 protein was examined in cultured DRG neurons (Fig. 6 A and B). Extensive observations were made in 12 separate cultures of ≈104 cells each. In cultures where neurite termini could be identified, PN1 immunoreactivity was observed predominantly in the growth cones, with lower levels of staining seen along the lengths of the neurites (Fig. 6B). All soma exhibited positive immunostaining for PN1, with modest variation in staining intensities among cells (Fig. 6A). The somal staining appeared to be confined to internal membranes because the staining was perinuclear and highly punctate, and was very similar in appearance to the staining pattern of the trans-Golgi network antigen TGN-38 in these cells (data not shown). In control experiments, preincubation of antibody with a 100-fold excess of the synthetic peptide antigen eliminated all of the immunofluorescence (except for a very weak, diffuse background fluorescence; data not shown).
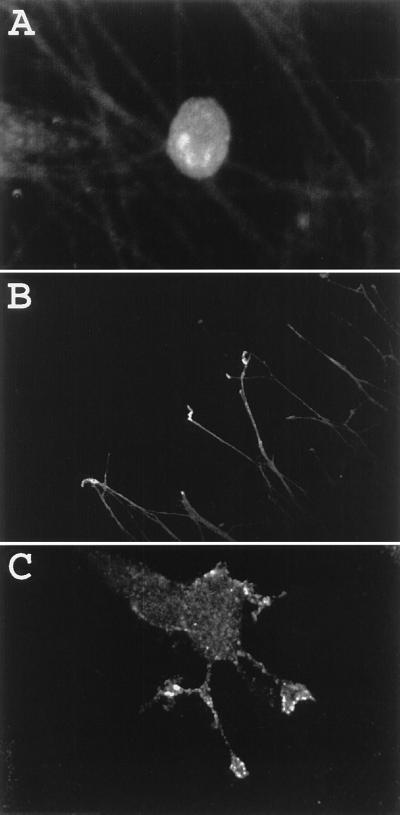
Figure 6.
Preferential targeting of PN1 to growth cone processes. Cultured E15 DRG neurons (A and B) and NGF-treated PC12 cells (C) were stained for PN1 epitopes using the PN1-specific antibody as described. (A and B, ×250; C, ×650.)
Because of previous studies indicating that PN1 transcripts were induced dramatically by treatment of PC12 cells with NGF, the distribution of PN1 protein in NGF-differentiated PC12 cells was also examined. Fused PC12 cells were NGF-treated for 2 days, a protocol sufficient for both induction of neurite growth (31) and PN1 sodium-channel expression (11, 18). Immunofluorescent localization of PN1 protein was carried out using the PN1-specific antibody as described. Specific staining of cells was never observed when the PN1 immunizing peptide was included with the antibody (data not shown). As seen in Fig. 6C, staining with the anti-PN1 antibody was highest at the neurite terminals, at the leading edges of the growth cones. Staining of growth cones was seen in all cells in which specific staining of PN1 protein was observed (>100 cells). Staining of cell bodies was occasionally observed at the cell periphery, particularly at sites of cell–cell contact (data not shown). Thus, the staining patterns in both cultured DRG neurons and NGF-treated PC12 cells are consistent with a targeting of PN1 protein from an intracellular pool to neurite terminals.
DISCUSSION
The molecular cloning experiments herein reveal that PN1 is a new member of the rat sodium channel gene family. Furthermore, this sodium channel type is expressed specifically, and to high levels, in neuronal populations throughout the PNS. The PN1 transcript corresponds to the mRNA induced selectively by brief treatment of PC12 cells with NGF (18), and likely corresponds to the sodium channel type whose existence in PNS was suggested from earlier RNA hybridization (10, 11) and immunochemical studies (12) using probes for conserved sodium channel sequences.
Surprisingly, comparison of the overall structure of PN1 to other known sodium channels indicates that it was likely the rat homolog (≈90% amino acid identity) of two recently identified human and rabbit sodium channels termed NeuroEndocrine sodium channel (hNE-Na; ref. 32) and Schwann cell sodium channel (Nas; ref. 33), respectively. The names of these two sodium channel types were assigned based upon isolation of their cDNAs from a neuroendocrine tumor cell line and cultured Schwann cells, respectively. In contrast to the Nas study, our concerted RNA hybridization and Western blot analyses indicate that the rat PN1 gene expression is restricted to neuronal populations within the PNS. A possible explanation for the apparent different tissue distributions is the use of RT-PCR in the Nas study, which is not a quantitative measure of the level of expression. Additionally, the Nas cDNA was cloned originally from cultured Schwann cells (33), and it is possible that genes are expressed aberrantly when these cells are placed in culture. Although the studies of both the human homolog of PN1 and rat PN1 revealed expression in adrenal gland, the former study did not examine for the presence of transcripts in the PNS, where PN1 is expressed to much higher levels. Thus, at this time, the most parsimonious explanation of all of the data is that the PN1/hNE–Na/Nas α subunit genes are expressed at the highest levels in PNS, and at much lower levels in brain, spinal cord, Schwann cells, and other tissues.
At least two sodium currents with different sensitivities to the sodium channel blocker TTX have been recorded from sensory neurons (13–15). In vertebrates, the proportions of the two currents correlate with the size of the neurons (14, 15), but the TTX-resistant sodium current is a minor component overall. The TTX-resistant sodium current is likely the result of expression of the newly described PN3/SNS α subunit, whose expression is restricted to small populations of sensory neurons in dorsal root, trigeminal, and nodose ganglia (16, 17). In contrast to the PN3/SNS sodium channel, the human homolog of PN1 (hNE-Na) gives rise to a TTX-sensitive sodium current when expressed in heterologous mammalian cells (32). We suggest that rat PN1, like its human homolog, is also TTX sensitive. In support of this idea, PN1 transcripts are expressed to high levels in two neuronal areas where TTX-resistant currents are not detected, superior cervical ganglion and medium and large diameter neurons within the DRG. Thus, these two distinct α subunit types (PN3/SNS and PN1/hNE–Na/Nas) may underlie two of the distinct sodium currents described previously in sensory neurons. It has been proposed that the PN3/SNS sodium channel participates in nociception. The presence of PN1 in the same ganglia as PN3/SNS, and the fact that NGF induces both PN1 mRNA (11, 18) and hyperalgesia (34–36), suggests the possibility that both sodium channel types may be involved in nociception.
The results from this study, combined with the results from two previous studies (11, 18), indicate that the PN1 sodium channel gene is induced by NGF in PC12 cells, concomitant with the appearance of membrane excitability. The previous electrophysiological studies were not able to determine the location of the sodium channels in the differentiated cells. Because of the generation of a PN1-specific antibody, we were now able to examine the subcellular distribution of PN1 protein in NGF-treated PC12 cells that had elaborated neuritic processes. These studies revealed the presence of PN1 protein in the neurite terminals. Furthermore, the distribution of PN1 α subunit in cultured DRG cells was similar to that observed in the PC12 cells. To our knowledge, this is the first immunocytochemical demonstration of the targeting of sodium channels to the terminal region, although previous studies have shown that different sodium channel types are targeted to distinct parts of neuronal cell membranes (37–42). Some staining of PN1 was detected in the cell bodies of both NGF-treated PC12 cells and the cultured DRG neurons. However, in this cellular compartment, very little PN1 protein appeared to be in the plasma membrane. The association of sodium channels with intracellular membranes has been noted previously for sodium channels in neurons, and may reflect pools of unassembled α subunits (42–46). Other ionic channels, including several potassium channels, are known to be targeted preferentially to the axon terminals (47–51). Although the mechanisms responsible for targeting of PN1 are not known, the localization of PN1 in the terminals suggest that this sodium channel is an additional important player in the final shaping of action potentials in the PNS.
Acknowledgments
We gratefully acknowledge William Therkauf for use of the confocal microscope. This work was supported by grants from the National Institutes of Health (NIH) (NS22518 and NS34375) and Trophix Pharmaceuticals to G.M., from NIH (NS15879 and NS34375) to S.R.L., and from NIH (NS18218 and NS34375) to S.H. J.J.T.-A. was supported by a Postdoctoral Fellowship from the National Multiple Sclerosis Society and B.L.M. was supported by a National Research Service Award.
Footnotes
Abbreviations: PNS, peripheral nervous system; PN1, peripheral nerve 1 sodium channel; DRG, dorsal root ganglia; NGF, nerve growth factor; TTX, tetrodotoxin; CNS, central nervous system; E, embryonic day.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U79568U79568).
References
- 1.Hille B. Ionic Channels of Excitable Membranes. 2nd Ed. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 2.Catterall W A. Physiol Rev. 1992;72:S15–S48. doi: 10.1152/physrev.1992.72.suppl_4.S15. [DOI] [PubMed] [Google Scholar]
- 3.Noda M, Ikeda T, Suzuki H, Takeshima H, Takahashi T, Kuno M, Numa S. Nature (London) 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- 4.Auld V J, Goldin A L, Krafte D S, Marshall J, Dunn J M, Catterall W A, Lester H A, Davidson N, Dunn R J. Neuron. 1988;1:449–461. doi: 10.1016/0896-6273(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 5.Kayano T, Noda M, Flockerzi V, Takahashi H, Numa S. FEBS Lett. 1988;228:187–194. doi: 10.1016/0014-5793(88)80614-8. [DOI] [PubMed] [Google Scholar]
- 6.Schaller K L, Krzemien D M, Yarowsky P J, Krueger B K, Caldwell J H. J Neurosci. 1995;15:3231–3242. doi: 10.1523/JNEUROSCI.15-05-03231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogart R, Cribbs L L, Muglia L K, Kephart D D, Kaiser M W. Proc Natl Acad Sci USA. 1989;86:8170–8174. doi: 10.1073/pnas.86.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trimmer J S, Cooperman S S, Tomiko S A, Zhou J, Cream S M, Boyle M B, Kallen R G, Sheng Z H, Barchi R L, Sigworth F G, Goodman R H, Agnew W S, Mandel G. Neuron. 1989;3:33–49. doi: 10.1016/0896-6273(89)90113-x. [DOI] [PubMed] [Google Scholar]
- 9.Kallen R G, Sheng Z-H, Yang J, Chen L, Rogart R B, Barchi R L. Neuron. 1990;4:233–242. doi: 10.1016/0896-6273(90)90098-z. [DOI] [PubMed] [Google Scholar]
- 10.Beckh S. FEBS Lett. 1990;262:317–322. doi: 10.1016/0014-5793(90)80218-8. [DOI] [PubMed] [Google Scholar]
- 11.D’Arcangelo G, Paradiso K, Shepherd D, Brehm P, Halegoua S, Mandel G. J Cell Biol. 1993;122:915–921. doi: 10.1083/jcb.122.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wollner D A, Catterall W A. Brain Res. 1985;331:145–149. doi: 10.1016/0006-8993(85)90724-3. [DOI] [PubMed] [Google Scholar]
- 13.Kostyuk P G, Velelovsky N S, Tsyndrenko A Y. Neuroscience. 1981;6:2423–2430. doi: 10.1016/0306-4522(81)90088-9. [DOI] [PubMed] [Google Scholar]
- 14.Campbell D T. Proc Natl Acad Sci USA. 1992;89:9569–9573. doi: 10.1073/pnas.89.20.9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy M L, Narahashi T. J Neurosci. 1992;12:2104–2111. doi: 10.1523/JNEUROSCI.12-06-02104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akopian A N, Sivilotti L, Wood J N. Nature (London) 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 17.Sangameswaran L, Delgado S G, Fish L M, Kock B D, Jakeman L D, Steward G R, Sze P, Hunter J C, Eglen R M, Herman R C. J Biol Chem. 1996;271:5953–5956. doi: 10.1074/jbc.271.11.5953. [DOI] [PubMed] [Google Scholar]
- 18.Toledo-Aral J J, Brehm P, Halegoua S, Mandel G. Neuron. 1995;14:607–611. doi: 10.1016/0896-6273(95)90317-8. [DOI] [PubMed] [Google Scholar]
- 19.Greene L A, Tischler A S. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobley W C, Scheuker A, Shooter E M. Biochemistry. 1976;15:5543–5552. doi: 10.1021/bi00670a019. [DOI] [PubMed] [Google Scholar]
- 21.Cathala G, Sabouret G S, Mendez B, West E L, Karin M, Martial J A, Baxter J D. DNA. 1983;2:329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- 22.Okamura Y, Ono F, Okagaki R, Chong J, Mandel G. Neuron. 1994;13:937–948. doi: 10.1016/0896-6273(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Proudfoot N J, Brownlee G G. Nature (London) 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 25.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 26.Cooperman S S, Grubman S A, Barchi R L, Goodman R H, Mandel G. Proc Natl Acad Sci USA. 1987;84:8721–8725. doi: 10.1073/pnas.84.23.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danielson P E, Forss-Petter S, Brau M A, Calavetta L, Douglass J, Milner R J, Sutcliffe J G. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 28.Fagan A M, Zhang H, Landis S, Smeyne R J, Silos-Santiago I, Barbacid M. J Neurosci. 1996;16:6208–6218. doi: 10.1523/JNEUROSCI.16-19-06208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dugandzija-Novakovic S, Koszowski A G, Levinson S R, Shrager P. J Neurosci. 1995;15:492–502. doi: 10.1523/JNEUROSCI.15-01-00492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp A H, Imagawa T, Leung A T, Campbell K P. J Biol Chem. 1987;262:12309–12315. [PubMed] [Google Scholar]
- 31.Halegoua S. Dev Biol. 1987;121:97–104. doi: 10.1016/0012-1606(87)90142-4. [DOI] [PubMed] [Google Scholar]
- 32.Klugbauer N, Lacinova L, Flockerzi V, Hofmann F. EMBO J. 1995;14:1084–1090. doi: 10.1002/j.1460-2075.1995.tb07091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belcher S M, Zerillo C A, Levenson R, Ritchie J M, Howe J R. Proc Natl Acad Sci USA. 1995;92:11034–11038. doi: 10.1073/pnas.92.24.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewin G R, Mendell L M. Trends Neurosci. 1993;16:353–359. doi: 10.1016/0166-2236(93)90092-z. [DOI] [PubMed] [Google Scholar]
- 35.Lewin G R, Ritter A M, Mendell L M. J Neurosci. 1993;13:2136–2148. doi: 10.1523/JNEUROSCI.13-05-02136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolf C J, Safieh-Garabedian B, Ma Q-P, Crilly P, Winter J. Neuroscience. 1994;62:327–331. doi: 10.1016/0306-4522(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 37.Ellisman M H, Levinson S R. Proc Natl Acad Sci USA. 1982;79:6707–6711. doi: 10.1073/pnas.79.21.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wollner D A, Catterall W A. Proc Natl Acad Sci USA. 1986;83:8424–8428. doi: 10.1073/pnas.83.21.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westenbroek R E, Merrick D K, Catterall W A. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- 40.Black J A, Friedman B, Waxman S G, Elmer L W, Angelides K J. Proc R Soc London Ser B. 1989;238:39–51. doi: 10.1098/rspb.1989.0065. [DOI] [PubMed] [Google Scholar]
- 41.Vabnick I, Novakovic S D, Levinson S R, Schachner M, Shrager P. J Neurosci. 1996;16:4914–4922. doi: 10.1523/JNEUROSCI.16-16-04914.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston W L, Dyer J R, Castellucci V F, Dunn R. J Neurosci. 1996;16:1730–1739. doi: 10.1523/JNEUROSCI.16-05-01730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt J, Rossie S, Catterall W A. Proc Natl Acad Sci USA. 1985;82:4847–4851. doi: 10.1073/pnas.82.14.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt J, Catterall W A. Cell. 1986;46:437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- 45.Brismar T, Gilly W F. Proc Natl Acad Sci USA. 1987;84:1459–1463. doi: 10.1073/pnas.84.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.French A S, Sanders E J, Duszyk E, Prasad S, Torkkeli P H, Haskins J, Murphy R A. J Neurobiol. 1993;24:939–948. doi: 10.1002/neu.480240707. [DOI] [PubMed] [Google Scholar]
- 47.Sheng M, Tsaur M-L, Jan Y N, Jan L Y. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 48.Sharma N, D’Arcangelo G, Kleinklaus A, Halegoua S, Trimmer J S. J Cell Biol. 1993;123:1835–1843. doi: 10.1083/jcb.123.6.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Kunkel D D, Schwartzkroin P A, Tempel B L. J Neurosci. 1994;14:4588–4599. doi: 10.1523/JNEUROSCI.14-08-04588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng M, Tsaur M-L, Jan Y N, Jan L Y. J Neurosci. 1994;14:2408–2417. doi: 10.1523/JNEUROSCI.14-04-02408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knaus H-G, Schwarzer C, Koch R O A, Eberhart A, Kaczorowski G J, Glossmann H, Wunder F, Pongs O, Garcia M L, Sperk G. J Neurosci. 1996;16:955–963. doi: 10.1523/JNEUROSCI.16-03-00955.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]