Abstract
Background
We previously identified eight known and novel genes differentially expressed in the small intestines of wild type and W/WV mice, which have greatly reduced populations of the interstitial cells of Cajal, that are responsible for the generation of electrical slow waves, by using a differential gene display method.
Methods
By using the same method we isolated additional candidate genes that were specifically down- or up-regulated in W/WV mice. Novel transcripts were designated as DDWMEST.
Results
We isolated seven candidates that were specifically down- or up-regulated in W/WV mice. Two novel transcripts, DDWMEST 1 and -91 were increased in both fed and fasted W/WV mice. Expression of another five genes was suppressed in W/WV mice: ARG2 (Arginase II), ONZIN (encoding leukemia inhibitory factor regulated protein), and three novel transcripts: DDWMEST62, -84, and -100. Together with the previous report, we identified fifteen differentially expressed genes in total in the small intestines of W/WV mice. Eight of these genes were reduced in the jejunums of W/WV mice compared to age matched wild type mice, whereas the other seven genes showed an increase in expression. Differential expression was the same in fasted and fed animals, suggesting that the differences were independent of the dietetic state of the animal.
Conclusions
Several known and novel genes are differentially expressed in the small intestines of W/WV mice. Differential gene comparison might contribute to our understanding of motility disorders associated with the loss of the interstitial cells of Cajal.
Background
Interstitial cells of Cajal (ICC) are pacemakers [1-3] and mediators of enteric motor neuro-transmission [4,5]. KIT, a receptor tyrosine kinase, is essential for the development and maintenance of the ICC phenotype [6,7]. Animals with reduced KIT expression (e.g the W/WV mutant mouse) develop few ICC at the level of the myenteric plexus (IC-MY) in the small intestine [2,3], and lose intramuscular ICC (IC-IM) in the stomach [4]. The loss of IC-MY is associated with a loss of electrical slow waves in the small intestine. Loss of IC-IM is associated with the disruption of enteric neuro-transmission in the gut [4].
Absolute or relative loss of ICC has been reported in many gastrointestinal (GI) motility disorders [8-12]. The association between diseases of GI motility and loss of ICC suggests that a more comprehensive understanding of the molecular biology of ICC may contribute to our understanding of the etiology of GI motility disorders [13-16]. The loss of IC-MY and slow wave activity in the small intestines of W/WV mice provides a unique opportunity to examine molecular changes that are associated with the loss of ICC in the gut. We hypothesized that there may be differential expression of genes in the small intestines of age-matched wild type and W/WV mice [15]. By the differential display method, we were able to identify a novel gene, ACPL1 and seven known genes that have been previously reported [15,16]. In both fasted and fed states, expression of the ACPL1, COX7B and SORCIN were considerably reduced in the jejunums of W/WV mice compared to age matched wild type mice. Whereas, five other genes (ADA, MDH1, RPL8, SPTB2, and p6-5) showed an increase in expression in fed and fasted W/WV mice Table 1.
Table 1.
Genes differentially expressed in the small intestine of W/WVmice
| Fragment Code | Accession No. | Protein (UniGene No.) | W/W V(a) | Status (b) | Subcellular location (c) | Chromosome (d) |
| DDWMEST117 | U82256 | ARG2 | decreased | this report | mitochondria(s) | 14q24.1-q24.3 |
| DDWMEST6 | Al263458 | ONZIN | decreased | this report | cytoplasm(p) | 4q21.22 |
| DDWMEST100 | W12272 | (Mm28954) | decreased | this report | cytoplasm(p) | 1q42.11-q42.3 |
| DDWMEST1 | AB052398 | unknown | increased | this report | ND | ND |
| DDWMEST62 | AB052399 | unknown | decreased | this report | ND | ND |
| DDWNEST84 | AB052400 | (Mm5296) | decreased | this report | nuclear(p) | 6 |
| DDWMEST91 | AB052402 | unknown | increased | this report | ND | ND |
| DDMNEST54 | AB030038 | ACPL1 | decreased | reported previously | cytoplasm(s) | 1q21 |
| DDWMEST101 | Al173010 | COX7B | decreased | reported previously | mitochondria(s) | Xq13.2 |
| DDWMEST4 | W33314 | SORCIN | decreased | reported previously | cytoplasm(s) | 7q21.1 |
| DDWMEST43 | U67771 | RPL8 | increased | reported previously | cytoplasm(s) | 8q24.3 |
| DDWMEST116 | M10319 | ADA | increased | reported previously | cytoplasm(p) | 20q12-q13.11 |
| DDWMEST90 | M16229 | MDH1 | increased | reported previously | cytoplasm(s) | 2p13.3 |
| DDWMEST110 | M74773 | SPTB2 | increased | reported previously | cytoplasm(s) | 2p21 |
| DDWMEST99 | M27347 | P6-5 | increased | reported previously | cytoplasm(p) | 12q13 |
(a) Gene expression pattern of each gene in the small intestine of W/WVmice. (b) "this report" means the gene newly isolated by us as a differentially expressed gene in the small intestine of W/WVmice. "reported previously" indicates that the differential expression of the gene was reported by us previously (ref.15, 16). (c) Subcellular location determined by the SOURCE program http://source.stanford.edu or predicted by the PSORTII program http://psort.ims.u-tokyo.ac.jp/form2.html were shown as "(s) " or " (p)", respectively. ND: not determined. Human chromosomal localization of the gene.
In the present study we have combined molecular profiles from an additional seven genes: We have identified fifteen candidate genes that are either up- or down-regulated in jejunal tissues of W/WV mice.
Methods
The use and treatment of experimental animals was approved by the guideline governing Animal Experiment Committee at the University of Yamanashi School of Medicine, Yamanashi, Japan and at the University of Nevada School of Medicine, Reno, USA. Six adult male WBB6F1-+/+ (wild type) and WBB6F1-W/WV mice, weighing 20 to 30 g, were purchased from Japan SLC Inc (Shizuoka, Japan). Animals were anesthetized with ether inhalation and sacrificed by cervical dislocation followed by exsanguation. Jejunal segments were resected and subsequently frozen in liquid nitrogen (-196°C).
For differential gene display, poly (A)+ RNA was isolated from each tissue sample by TRIZOL (Life Technologies, Inc., Gaithersburg, MD, USA). The differential display procedure was performed as previously described [15-18]. The blots were autoradiographed and analyzed with a fluorescent image analyzer (FMBIO, Takara Co., Kyoto, Japan). For reverse transcription-polymerase chain reaction (RT-PCR), total RNA was isolated from resected specimens by TRIZOL. cDNA were prepared from 2 μg of total RNA by (dT)15 priming and synthesized in the same manner as in the differential gene display experiments. The levels of gene expression were analyzed by semi-quantitative RT-PCR using mRNAs from proximal jejunums of fed and fasted wild type and W/WV mice as templates. The PCR reactions were optimized for number of cycles to ensure product intensity within the linear phase of amplification.
Results
To identify genes that may be selectively associated with IC-MY, we compared gene expression patterns in jejunal tissues derived from age-matched wild type and W/WV mice utilizing a differential gene display method. Using various primer sets, we identified a total of twelve cDNA bands that were likely to be expressed at the biggest different levels in these tissues. They showed either intense or weak labeling in the jejunums of wild type animals but very weak or intense labeling in W/WV jejunums. To confirm the differential display profiles, we further examined the expression levels in murine jejunal mRNA derived from wild type and W/WV mice maintained under fed or fasted conditions as templates, using semi-quantitative RT-PCR analysis. In this way, we identified a total of eight differentially expressed genes in the small intestines of W/WV mice Table 1.
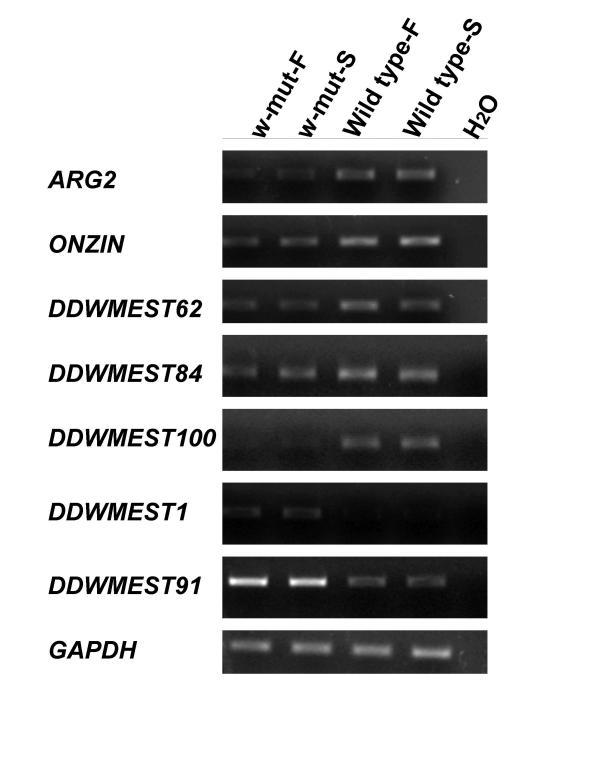
We identified a total of nine cDNA bands expressed at the biggest different levels in the small intestines of W/WV mice by using primer sets, which were not tested in the previous study. By means of semi-quantitative RT-PCR analysis with subsequent cDNA screening, we were able to isolate seven additional novel candidate transcripts among the nine bands that were specifically down- or up-regulated in the small intestines of W/WV mice; two of them (ARG2 and ONZIN) were found to be known genes (Table 1, Fig 1). Semi-quantitative RT-PCR revealed that the other five candidates (DDWMEST-1, -62, -84, -91, and -100; tentative names) were differentially expressed in W/WV mice (Table 1, Fig. 1), although in some of them full-length cDNA have not been obtained so far. A search with the FASTA program http://www.ebi.ac.uk/fasta33/ revealed no significant homology between these 5 novel transcripts and any archived molecule in the public databases, however two of them, DDWMEST84 and -100 were found in the UniGene Cluster database that contains sequences that represent a unique gene, as well as related information such as the tissue types in which the gene has been expressed and genetic map location http://www.ncbi.nlm.nih.gov/UniGene/[19]. In both fasted and fed states, expression of the DDWMEST 1 and -91 were significantly increased in the jejunums of W/WV mice compared to age matched wild type mice. Whereas, the other five transcripts (ARG2, ONZIN, DDWMEST-62, -84, and -100) showed a decrease in expression in fed and fasted W/WV mice.
Figure 1.
Expression of seven genes, using semi-quantitative RT-PCR analysis, in jejunum tissues from wildtype and W/WV mutant mice maintained under fed and fasted conditions. F and S indicate samples of fed and starved mice respectively. GAPDH levels were measured as a control. Five genes showed a decrease in expression in both fed and fasted W/WV mice. These transcripts included: the gene for ARG2, ONZIN, DDWMEST62, -84, and -100. Two transcripts, DDWMEST1 and -91 were increased in both fed and fasted W/WV mice.
Discussion
In the present study we have identified seven additional differentially expressed genes in the small intestines of W/WV mice. This animal model have the loss of IC-MY in the gut that are responsible for the generation of electrical slow wave activity. In combination with our previous reports [15,16], now we have completed the differential display analysis and all the twenty-one candidate cDNA bands that were likely to be expressed at the reliable different levels in those tissues confirmed with semi-quantitative RT-PCR, the differential expression of a total of fifteen genes, and obtained a molecular profile of the small intestine of W/WV mice. A comparison of differentially expressed genes from the jejunums of W/WV mice using a differential display method has revealed that the expression of eight genes were significantly suppressed in W/WV mice, whereas seven genes were dramatically up-regulated in these animals Table 1. Data obtained from these experiments suggest that expression of the genes were specifically regulated in W/WV mice and were independent of the dietary conditions (i.e. fed or fasted) imposed.
The two additional genes that were up-regulated in the small intestines of W/WV mice were identified; DDWMEST-1 and -91, both of which revealed no significant homology to any known genes. Two of five additional down-regulated genes in W/WV mice, ARG2 and ONZIN were identified as known genes by cDNA screening. ARG2 has seven conserved residues known to be essential for activity of arginases [20], and has an important role in the regulation of nitric oxide synthesis and polyamine metabolism by modulating local arginine concentrations [21]. It also contains a mitochondrial import sequence, and is imported into the mitochondria with appropriate proteolysis [22]. It is reported that release of cytochrome c from the mitochondrial intermembrane space results in nuclear apoptosis [23]. An abundance of mitochondria is one of the criteria used to identify IC-MY by electron microscopy, and our data have possibly suggested the importance of mitochondria in generating pacemaker activity in GI muscles [24]. The loss of IC-MY in W/WV mutant mice may be associated with the down-regulation of COX7B and ARG2 in these animals, both of which were localized in the mitochondria. ONZIN was identified through a screening of LIF regulated genes. LIF is a multifunctional cytokine implicated in various functions and is supported to play a role in neuro-immune reactions [25] as well as an important role in aspects of neuronal development [26,27]. Expression of LIF protein is observed in the gut, especially in the myenteric and submucous ganglion cells at 13–31 weeks of gestation in childhood cases and adults [28]. The down-regulation of ONZIN in W/WV mice therefore suggests the involvement of the LIF pathway in the GI motility disorders.
The other three transcripts whose expression were decreased in W/WV mice showed no significant homology to any known genes, however two of them, DDWMEST-84 and -100, matched to the assembled sequences in the UniGene Cluster database.19 DDWMEST-84 (HomoloGene No.: Mm.5296) was reported to be expressed in brain, embryo, heart, lung, and mammary gland and to be mapped to mouse chromosome 10, and its human counterpart was localized at chromosome 6. DDWMEST-100 (HomoloGene No.: Mm.28954) was suspected to be expressed ubiquitously in various tissues, such as embryo, hypothalamus, liver, muscle, placenta, skin, lung, mammary gland, and uterus.
In the fifteen genes identified, we also determined the subcellular localization and human chromosomal mapping of the twelve known genes and two UniGene Cluster sequences using several web-available software (the SOURCE program: http://source.stanford.edu or the PSORTII program: http://psort.ims.u-tokyo.ac.jp/form2.html Table 1. Interestingly, eleven genes were suggested to be localized in the cytoplasm or mitochondria. Another gene was supposed to encode a nuclear protein. Further analyses of these genes might enable us to elucidate not only their relationship with the KIT, a receptor tyrosine kinase, but also the molecular aspects of GI pacemaker system.
Today the Genome Project is considered to be one of the most important projects in biology and medicine. The discovery of the entire human and mouse genes through this project will revolutionize biological medicine including molecular diagnosis of various diseases and the development of novel treatments. The information will accelerate discovery of genes susceptible to or causing various diseases and contribute to screening of novel drugs that target these disease-gene products. In this regard, the effort of the molecular profiling project in which we attempt to discover genetic aberrations in animal disease models such as W/WV mice will generate very variable resources for further elucidation of GI motility disorders.
At the present time we do not have enough evidence whether these genes are important to the function of the pacemaker apparatus or were down-or up-regulated as a result of the loss of ICC and impairment of normal slow wave pacemaker activity. However, the molecular profile obtained can yield valuable insights into the molecular events underlying motility disorders in which ICC are lost. Application of the differential gene display method, when combined with the future progress of the human and mouse genome project, may reveal new genetic pathways that are common to those GI motility disorders characterized by a deficiency of ICC.
Conclusion
Our data provide one of the first genetic evidences that several important proteins that have roles in DNA/RNA/protein synthesis and metabolism, oxygen metabolism, energy cycle, and development and differentiation of gut cells are significantly changed in the small intestines of W/WV mice. Considering the number of novel genes identified by us in this study, we suggest that many unknown genes could be involved in the cellular changes that lead to motility disorders associated with ICC loss.
List of abbreviations
ACPL1: acid phosphatase-like protein 1,
ADA: adenosine deaminase,
ARG2: Arginase II,
COX7B: cytochrome c oxidase subunit VIIb,
DMP: deep muscular plexus,
ESTs: expressed sequence tags,
FISH: fluorescence in situ hybridization,
GI: gastrointestinal,
GAPDH: glyceraldehyde-3-phosphate dehydrogenase,
ICC: interstitial cells of Cajal,
IC-DMP: ICC at the level of the deep muscular plexus,
IC-IM: intramuscular ICC,
IC-MY: ICC at the level of the myenteric plexus,
LIF: leukemia inhibitory factor,
LOS: lower esophageal sphincter,
MY: myenteric plexus,
MDH1: malate dehydrogenase,
SPTB2: spectrin, nonerythroid, beta subunit,
RPL8: ribosomal protein L8,
RT-PCR: reverse transcription-polymerase chain reaction.
Competing interests
NONE DECLARED.
Authors' contributions
YD and IT carried out the molecular studies and the design of study and coordination. BAJP and CC participated in the molecular studies. SW, KS and MF participated in the design of study and coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
Supported by 08457165 (to MF) from the Japanese Ministry of Education, Science, Sports and Culture, JAPAN and DK57236 (to SW) and DK41315 (to SW and KS) from the National Institutes of Health, USA.
Contributor Information
Yataro Daigo, Email: yd206@cam.ac.uk.
Ichiro Takayama, Email: ichirot@is.icc.u-tokai.ac.jp.
Bruce AJ Ponder, Email: bajp@mole.bio.cam.ac.uk.
Carlos Caldas, Email: cc234@cam.ac.uk.
Sean M Ward, Email: sean@physio.unr.edu.
Kenton M Sanders, Email: kent@physio.unr.edu.
Masayuki A Fujino, Email: mfujino@res.yamanashi-med.ac.jp.
References
- Langton PD, Ward SM, Carl A, Norell MA, Sanders KM. Spontaneous electrical activity of interstitial cells of Cajal isolated from canine proximal colon. Proc Natl Acad Sci USA. 1989;86:7280–7284. doi: 10.1073/pnas.86.18.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Burns AJ, Torihashi S, Sanders KM. Mutation of the proto-oncogene c-kit blocks development of interstitial cells and electrical rhythmicity in murine intestine. J Physiol (Lond) 1994;480:91–97. doi: 10.1113/jphysiol.1994.sp020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizinga JD, Thuneberg L, Kluppel M, Malysz J, Mikkelesen HB, Bernstein A. W/kit gene required for interstitial cells of Cajal and for intestinal pacemaker activity. Nature (Lond) 1995;373:347–349. doi: 10.1038/373347a0. [DOI] [PubMed] [Google Scholar]
- Burns AJ, Herbert TM, Ward SM, Sanders KM. Interstitial cells of Cajal inhibit neurotransmission in the stomach. Proc Natl Acad Sci USA. 1996;93:12008–12013. doi: 10.1073/pnas.93.21.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Beckett EAH, Wang XY, Baker F, Khoyi M, Sanders KM. Interstitial cells of Cajal mediate enteric excitatory neurotransmission in the murine fundus. J Neurosci. 2000;20:1393–1403. doi: 10.1523/JNEUROSCI.20-04-01393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Tokutomi N, Sato D, Tokutomi Y, Sugita M, Lai ZF, Torihashi S. Identification of gastrointestinal tract pacemaker cells and their functional significance. In: Tsuchiya M, Matsuo Y, Kasuga Y, Muto T, editor. Gastrointestinal Function, Regulation and Disturbance. Vol. 14. Tokyo: Excepta Medica; 1996. pp. 27–43. [Google Scholar]
- Takayama I, Takeda M, Ohno S, Fujino MA. Experimental pacemaker disease model in mouse jejunum: Impairment of interstitial cells of Cajal by an antagonistic antibody for c-kit. Yamanashi Med J. 1998;13:53–63. [Google Scholar]
- Isozaki K, Hirota S, Miyagawa J, Taniguchi M, Shiomura Y, Matsuzawa Y. Deficiency of c-kit + cells in patients with a myopathic form of chronic idiopathic intestinal pseudo-obstruction. Am J Gastroenterol. 1997;92:332–334. [PubMed] [Google Scholar]
- Vanderwinden JM, Liu H, DeLaet NH, Vanderhagen JJ. Interstitial cells of Cajal in human colon and in Hirschsprung's disease. Gastroenterology. 1996;111:901–910. doi: 10.1016/s0016-5085(96)70057-4. [DOI] [PubMed] [Google Scholar]
- He CL, Burgart L, Wang L, Pemberton J, Young-Fadok L, Szurszewski J, Farrugia G. Decreased interstitial cell of Cajal volume in patients with slow-transit constipation. Gastroenterology. 2000;118:14–21. doi: 10.1016/s0016-5085(00)70409-4. [DOI] [PubMed] [Google Scholar]
- Ordog T, Takayama I, Cheung WKT, Ward SM, Sanders KM. Remodeling of networks of interstitial cells of Cajal in a murine model of diabetic gastroparesis. Diabetes. 2000;49:1731–1739. doi: 10.2337/diabetes.49.10.1731. [DOI] [PubMed] [Google Scholar]
- Takayama I, Seto E, Zai H, Ohno S, Tezuka H, Daigo Y, Fujino MA. Changes of in vivo gastroenteric motor pattern in pacemaker-deficient (WsRC-Ws/Ws) rats. Dig Dis Sci. 2000;45:1901–1906. doi: 10.1023/A:1005612109863. [DOI] [PubMed] [Google Scholar]
- Takayama I, Daigo Y, Kojima Y, Fujino MA. Gastroenteric pacemaker system. Nippon Shoukakibyo Gakkai Zasshi. 2001;98:922–934. [PubMed] [Google Scholar]
- Takayama I, Horiguchi K, Daigo Y, Mine T, Fujino MA, Ohno S. The interstitial cells of Cajal and a gastrointestinal pacemaker system. Arch Histol Cytol. 2002;65:1–26. doi: 10.1679/aohc.65.1. [DOI] [PubMed] [Google Scholar]
- Takayama I, Daigo Y, Ward SM, Sanders KM, Yamanaka T, Fujino MA. Differential gene expression in the small intestines of wild type and W/W V mice. Neurogastroenterol Motil. 2001;13:163–168. doi: 10.1046/j.1365-2982.2001.00256.x. [DOI] [PubMed] [Google Scholar]
- Takayama I, Daigo Y, Ward SM, Sanders KM, Walker RL, Horowitz B, Fujino MA. Novel human and mouse genes encoding an acid phosphatase family member and its down regulation in W/WV mouse jejunum. Gut. 2002;50:790–796. doi: 10.1136/gut.50.6.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display. A general protocol. Mol Biotechnol. 1998;10:261–267. doi: 10.1007/BF02740847. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, et al. A gene map of the human genome. Science. 1996;274:540–546. doi: 10.1126/science.274.5287.540. [DOI] [PubMed] [Google Scholar]
- Morris SM, Jr, Bhamidipati D, Kepka-Lenhart D. Human type II arginase: sequence analysis and tissue-specific expression. Gene. 1997;193:157–161. doi: 10.1016/S0378-1119(97)00099-1. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Sonoki T, Nagasaki A, Terada K, Takiguchi M, Mori M. Molecular cloning of cDNA for no hepatic mitochondrial arginase (arginase II) and comparison of its induction with nitric oxide synthase in a murine macrophage-like cell line. FEBS Lett. 1996;395:119–122. doi: 10.1016/0014-5793(96)01015-0. [DOI] [PubMed] [Google Scholar]
- Vockley JG, Jenkinson CP, Shukla H, Kern RM, Grody WW, Cederbaum SD. Cloning and characterization of the human type II arginase gene. Genomics. 1996;38:118–123. doi: 10.1006/geno.1996.0606. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Ward SM, Ordog T, Koh SD, Abu Baker S, Jun JY, Amberg G, Sanders KM. Pacemaking in interstitial cells of Cajal depends upon calcium handling by endoplasmic reticulum and mitochondria. J Physiol (Lond) 2000;525:355–361. doi: 10.1111/j.1469-7793.2000.t01-1-00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint VB, Briere F, Bates EE. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- Escary JL, Perreau J, Dumenil D, Ezine S, Brulet P. Leukaemia inhibitory factor is necessary for maintenance of haematopoietic stem cells and thymocyte stimulation. Nature. 1993;363:361–364. doi: 10.1038/363361a0. [DOI] [PubMed] [Google Scholar]
- Koblar SA, Turnley AM, Classon BJ, Reid KL, Ware CB, Cheema SS, Murphy M, Bartlett PF. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc Natl Acad Sci USA. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester T, Olsen L. Expression of leukaemia inhibitory factor during the development of the human enteric nervous system. Histochem J. 2000;32:345–334. doi: 10.1023/A:1004061529723. [DOI] [PubMed] [Google Scholar]