Abstract
Ran-binding protein 2 (RanBP2) (type II) is a retinal cyclophilin-related protein that binds Ran-GTPase. Type I cyclophilin is a shorter, alternatively spliced isoform of RanBP2. Recently, we showed that the Ran-binding domain 4 (RBD4)/cyclophilin (CY) supradomain of RanBP2 acts both in vitro and in vivo as a specific chaperone for bovine red/green opsin (R/G opsin). R/G opsin undergoes a stable modification of its electrophoretic mobility upon binding to RanBP2. This modification is likely due to cis–trans isomerization of one or more proline residues in the opsin protein. Here, we show that expression of human red opsin in Escherichia coli and COS cells results in the production of still a third electrophoretic variant of this protein. This variant was converted to the RBD4 binding-competent form of opsin through direct interaction with RBD4/CY, both in vivo and in vitro. We suggest that these distinct opsin species may represent kinetically or thermodynamically trapped prolyl conformers that can be interconverted by concerted action of the RBD4 and CY domains of RanBP2. We also show that the C-terminal half of RBD4 is the binding domain for bovine R/G opsin and that coexpression of human red opsin with type I cyclophilin in vivo enhances the production of functional visual pigment. These observations imply that prolyl isomerization may have importance beyond its role in protein folding, possibly as a molecular switch modulated by cyclophilin for the loading of opsin onto RanBP2 during visual pigment processing in cones.
Keywords: peptidyl-prolyl cis–trans isomerase, Ran-GTPase, biogenesis
Peptidyl-prolyl cis–trans isomerases (PPIases) catalyze the cis–trans isomerization of prolyl-peptide bonds (1–6), a rate-limiting step in protein folding (7–9). Some PPIases also may possess chaperone activity by binding and inhibiting the formation of misfolded protein aggregates (10–12). It remains unclear if the PPIase and chaperone activities are independent functions of these proteins (6, 13–15). There is emerging evidence implicating PPIase proteins in modulation of channel and receptor activities (14, 16–18), morphogenesis of HIV-1 virions (19), regulation of mitosis (15), and processing, transport and/or maturation of transmembrane receptors (20–22). PPIases are ubiquitous and abundant proteins, divided into four unrelated subgroups: the cyclophilins (1–4, 23), the FK506-binding proteins (1–4), the parvulins (5, 24, 25), and the trigger factor (6, 26). Cyclophilins comprise a large class of conserved proteins that bind the potent immunosuppressive drug cyclosporine A (23, 27). Until recently, the only cyclophilin with a suggested function in vivo was the NinaA protein of Drosophila. Mutations in the ninaA gene severely reduce the levels of opsins only in a subclass of fly photoreceptors (21, 22, 28–30). In addition, the NinaA protein forms a stable complex in vivo with R1–6 opsin (31). NinaA thus plays a critical role in opsin biogenesis. The molecular mechanisms underlying the function and opsin subclass specificity of NinaA, however, are not understood.
In an attempt to identify proteins in mammalian retina with biological properties similar to those of NinaA, we isolated splice variants of a new class of cyclophilin-related proteins, types I and II (32, 33). These are expressed predominantly in cones among photoreceptor cells (33). The type II isoform is identical to Ran-binding protein 2 (RanBP2) (34, 35), a large protein that contains tandem domains homologous to Ran-binding protein 1 (RanBP1) (36) and binds Ran GTPase (37). To investigate the role of RanBP2 in retinal function, we recently analyzed the function of two adjacent domains of RanBP2, Ran-binding domain 4 (RBD4), and the cyclophilin domain (CY), in the presence of bovine retinal extracts (38). We showed that the RBD4/CY supradomain forms a complex with bovine red/green opsin (R/G opsin) but not with the closely related blue-cone or rod opsins. The CY does not itself bind R/G opsin but stabilizes, both in cis and trans, formation of the R/G opsin-RBD4 complex. This chaperone function is associated with a CY-mediated modification of R/G opsin, reflected by a shift in the predominant electrophoretic mobility of opsin from 49.5 to 51 kDa. This modification is dependent on the CY-PPIase activity, and thus may be due to cis–trans isomerization of one or more proline residues within opsin. Coexpression of human red opsin and RBD4/CY in COS cells increased the production of functional visual pigment, suggesting that the specific chaperone and “foldase” activity of RanBD4/CY is important in the processing and maturation of human red opsin. To further investigate the effect of RBD4/CY and type I cyclophilin on R/G opsin, we used a heterologous coexpression system to carry out extended structure–function analysis on RBD4 and CY in bovine retinal extracts.
MATERIALS AND METHODS
General Methods.
SDS/10% polyacrylamide gels (1.5 mm thick, 13 × 13 cm) were run on a Hoefer SE600 electrophoresis apparatus. Blotting of SDS/polyacrylamide gels onto polyvinylidene difluoride (Immobilon-P) membranes (Millipore) was carried out on a Genie electrophoretic blotter (Idea Scientific, Corvallis, OR). Crude bovine retinal extracts were prepared by grinding 10 bovine retinas (Pel-Freez Biologicals) to fine powder on dry ice followed by homogenization in a glass homogenizer with 30 ml of retinal homogenization buffer {0.75% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate]/20 mM Tris·HCl, pH 6.8/250 mM NaCl/2 mM 2-mercaptoethanol/0.02% NaN3/5% (vol/vol) glycerol and protease inhibitors}. Retinal homogenates were centrifuged and precleared with 5 ml of swollen glutathione S-agarose beads (Sigma) and 500 μg of glutathione S-transferase (GST) (Sigma) for about 30–60 min at 4°C. Protein was quantitated by the Bio-Rad protein assay.
Preparation, Expression, and Purification of Recombinant Proteins.
GST-fused human red opsin construct was prepared by subcloning in-frame the NcoI–HindIII fragment of clone hs7 (39) into the bacterial expression vector pGEX-KG (40), followed by transformation into Escherichia coli strain XL1-Blue (Stratagene). Expression of GST-fused human red opsin was induced with isopropyl 1-thio-β-d-galactopyranoside (1 mM) for 3–4 h. Cells were pelleted, resuspended in retinal homogenization buffer, and lysed twice in a French pressure cell followed by precipitation of the cell debris. Affinity purification of red opsin was carried out by loading the bacterial lysate onto a column (Bio-Rad) with swollen glutathione-S-agarose beads and incubating for 1 h at 4°C. The eluate was discarded, and beads were incubated and washed four times with 15 ml of washing buffer (50 mM Tris·HCl, pH 7.5/100 mM NaCl/2 mM CaCl2/2 mM MgCl2/0.2% Triton X-100) followed by a washing step with thrombin cleavage buffer at 25°C (50 mM Tris·HCl, pH 8.0/150 mM NaCl/2.5 mM CaCl2/0.1% 2-mercaptoethanol/0.2% Triton X-100). Human red opsin was cleaved from GST by incubating the agarose beads with thrombin cleavage buffer and thrombin (Sigma). All other GST-fused proteins (GST-RBD4/CY and fragments thereof) were constructed and expressed exactly as previously described (33, 38, 40). Proteins were concentrated and washed two or three times using Centricon concentrators (Amicon) (38). Expression of unfused human red opsin used for coexpression in E. coli with GST-RBD4/CY was carried out using the bacterial expression vector pALTER-Ex2 (Promega) with the start translation site of red opsin placed eight bases downstream from the ribosome-binding site. pALTER-Ex2 is an expression vector driven by the tac promoter containing a tetracycline selective marker and p15a origin of replication compatible with ColE1 vectors such as pGEX-KG (40). The human red opsin clone hs7 was isolated by digestion with NcoI, partial filling-in of the NcoI-sticky end with the Klenow fragment of DNA polymerase, dCTP, dATP, and dTTP followed by EcoRI digestion. The pALTER-Ex2 vector was digested with BamHI followed by partial filling of the BamHI-sticky end with the Klenow fragment, dGTP, dATP, dTTP, and EcoRI digestion. The resulting fragments were ligated, mixed with GST-RBD4/CY plasmid, and coelectroporated into the laqIq host E. coli strain, NM522.
In Vitro and in Vivo Binding and Electrophoretic Mobility-Shift Assays.
Binding assays in bovine retinal extracts with GST-fused proteins were carried out at 4°C and 26°C, exactly as previously described (38). Electrophoretic mobility-shift assays in vitro with human red opsin expressed and purified from E. coli and COS cells were performed by mixing and incubating at 4°C equimolar concentrations (2 μM) of human red opsin and RBD4/CY, RBD4, or CY in incubation buffer (50 mM Tris·HCl, pH 7.5/100 mM NaCl/2 mM CaCl2/2 mM MgCl2/0.5% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate) for 1 h. Reactions were stopped with the addition of 1 vol of 2× SDS/PAGE sample buffer and boiling of the samples for 3–5 min. For in vivo binding analysis of GST-RBD4/CY to unfused human red opsin, NM522 cells were coelectroporated with the respective constructs. Protein expression was induced with isopropyl l-thio-β-d-galactopyranoside (1 mM) for about 3 h, and the cells were pelleted and resuspended in 8:3 retinal homogenization/incubation buffer, lysed twice in a French pressure cell, followed by centrifugation of cell debris. The GST-RBD4/CY-red opsin complex was purified by incubating the lysate supernatant with glutathione-agarose beads (Sigma), precipitating and washing of the beads three times with washing buffer followed by elution of the GST-complex with 10 mM reduced glutathione (Sigma) in washing buffer. Aliquots were boiled after the addition of 2× SDS/PAGE sample buffer. Western blot analysis of incubation reactions was carried out with rabbit anti-human red opsin (JH 492, 1:2500) (41) using a chemiluminescent substrate (Kirkegaard & Perry Laboratories) as described (38).
Characterization of the Effects of Type I Cyclophilin on Human Red Pigment Generation.
For the expression of type I cyclophilin with two different start sites (33), the corresponding cDNAs were subcloned into EcoRI and NotI sites of the expression vector pMT3. The transient expression of red opsin with and without type I cyclophilin in COS-1 cells, pigment generation with 11-cis-retinal, purification of red pigment, and spectroscopic analysis of the purified pigment were according to the procedures previously described (38).
RESULTS
In Vitro Binding Analysis of RBD4/CY to Red Opsin.
To dissect further the role of RBD4/CY in R/G opsin binding, we made a series of RBD4/CY-fused constructs (Fig. 1), incubated them in parallel with detergent-solubilized retinal extracts at 4°C and room temperature, and analyzed the coprecipitating substrates by Western blotting with an antibody against red opsin (Fig. 2). Previously, we showed that deletion of CY from the RBD4/CY supradomain led to a significant reduction in the binding of opsin to RBD4 when binding reactions were carried out at 4°C (38). In addition, removal of the N-terminal half of RBD4 from RBD4/CY (equivalent to type I cyclophilin) abolished the binding of opsin to RBD4. The N-terminal half of RBD4 (N-RBD4) by itself, however, did not bind R/G opsin. Interestingly, when we incubated the C-terminal half of RBD4 (RBD4-C) without CY, we saw significant binding of opsin at levels comparable to that observed with the whole RBD4 domain (Fig. 2). In addition, incubation of binding reactions in parallel at a higher temperature led to a significant increase in the binding affinity of red opsin to RBD4-C/CY (equivalent to type I cyclophilin) (Fig. 2 Right, lane 3) without changing the affinities of binding of opsin to any of the other constructs tested. This interaction appears to be specific, because we did not observe any nonspecific binding of opsin to the N-terminal half of RBD4 and CY.
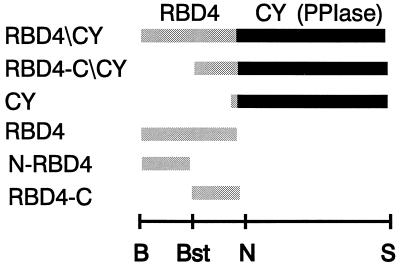
Figure 1.
Schematic diagram of six bovine RBD4/CY recombinant constructs used in this study. RBD4/CY contains the entire RBD4 and CY with PPIase catalytic site. RBD4-C/CY contains the C-terminal half of RBD4 and the whole CY. This construct represents type I cyclophilin, assuming the second methionine is used for translation initiation (33). CY contains only the cyclophilin domain. RBD4 contains the entire RBD4. N-RBD4 contains the N-terminal half of RBD4. RBD4-C contains the C-terminal half of RBD4. The restriction sites of the cognate cDNA used for the cloning of the RanBP2 domains into the GST-expression vector, pGEX-KG, are shown below the constructs. B, BamHI; Bst, BstXI; N, NcoI; S, StuI.
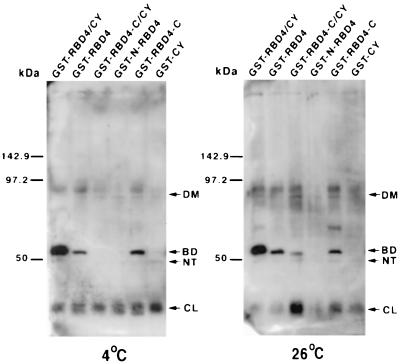
Figure 2.
The C-terminal half of RBD4 is the binding domain for bovine R/G opsin. Western blot analysis of glutathione-S-agarose coprecipitates from incubation reactions of bovine retinal extracts with GST-RBD4/CY and subfragments thereof (Fig. 1) at 4°C (Left) and 26°C (Right) using an antibody against human red opsin (41). Removal of the N-terminal half of RBD4 from RBD4/CY (RBD4-C/CY) abolishes its binding to R/G opsin at 4°C. This binding could be restored by further removal of CY from RBD4-C/CY (RBD4-C). The increase of the incubation temperature from 4°C to 26°C made RBD4-C/CY competent to bind R/G opsin without changing the affinity of R/G opsin to any other constructs. CL, NT, BD, and DM represent, respectively, the 34-kDa collapsed, traces of the 49.5-kDa native, 51-kDa RBD4/CY-binding competent form, and traces of red opsin dimers.
In Vivo Role of Type I Cyclophilin (RBD4-C/CY).
The RBD4-C/CY construct encodes a protein identical to type I cyclophilin (33). Previously, we observed that coexpression of RBD4 and CY led to an increase in the production of functional pigment. To understand further the role of type I cyclophilin (RBD4-C/CY) on red opsin, we investigated its function in vivo. We singly expressed red opsin or coexpressed it with type I cyclophilin or RBD4/CY in COS cells. Because there is ambiguity about which of two AUG codons functions as the type I cyclophilin translation-initiator, we assembled two constructs for type I cyclophilin, one encoding a protein with an additional 17 residues at the N terminus. Compared with cells expressing red opsin alone, coexpression of red opsin with RBD4/CY or with either form of type I cyclophilin led to similar increases in the formation of functional visual pigment, with little change in the production of opsin apoprotein (Fig. 3).
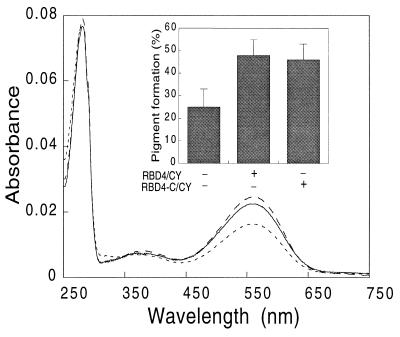
Figure 3.
UV–visible absorption spectra of red pigment expressed in COS cells. Red opsin expressed with RBD4-C/CY (type I cyclophilin) (solid line), with RBD4-CY (dotted line), and alone (broken line) was reconstituted with 11-cis-retinal and purified as described in the text. Coexpression of red opsin with type I cyclophilin [the second starting methionine (33) was used as translational start site] increased pigment generation similar to that as with RBD4-CY (Inset). Similar results were obtained when the first starting methionine of type I cyclophilin was used as a translational start site (data not shown). The spectra were normalized to the same scale at 280 nm absorbance. The typical absolute 280 values ranged from 0.07 to 0.8 for red opsin alone, from 0.6 to 0.8 for red opsin with type I cyclophilin, and from 0.5 to 0.7 for red opsin with RBD4-CY.
Interconversion of Red Opsin Isoforms in Vivo and in Vitro Directly by RBD4/CY.
Next, we studied the effect of red opsin expression, singly or in combination with RBD4/CY, in E. coli cells. To this end, we first subcloned the human red opsin cDNA into pALTER-Ex2 vector (Promega). Transformation of the unfused opsin expression construct into strains DH-5α and lacIq host, NM522, resulted in no clones or only nonrecombinant clones, respectively. A possible explanation is cytotoxicity caused by leaky expression of unfused opsin. To overcome this potential toxic effect, we either expressed red opsin fused to GST in NM522 cells or coexpressed unfused opsin with GST-RBD4/CY. Both approaches resulted in the growth of viable recombinants. Affinity purification of the fused red opsin, followed by proteolytic cleavage of opsin from GST, and Western blot analysis with an antibody against red opsin, revealed the production of a major 65-kDa opsin isoform (Fig. 4A, lane 1). In contrast, cotransformation of NM522 cells with GST-fused RBD4/CY and unfused opsin constructs resulted in the expression of a 51-kDa RBD4/CY-binding-competent form of red opsin (Fig. 4A, lane 2).
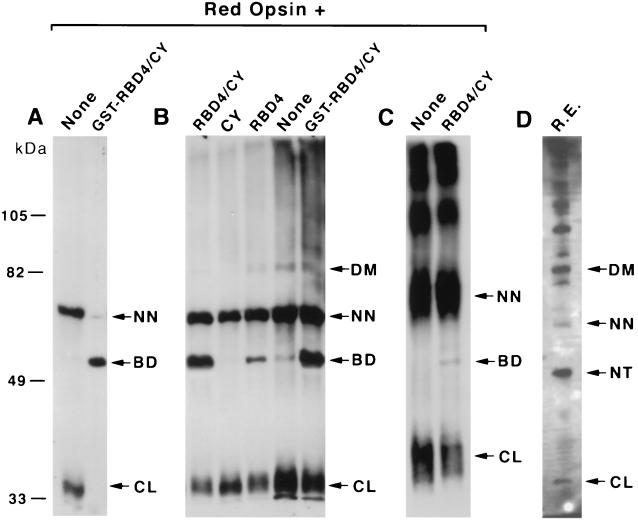
Figure 4.
In vivo and in vitro binding and electrophoretic mobility-shift assays of RBD4/CY, RBD4, and CY on red opsin. (A) Western blot of red opsin expressed by itself or coexpressed with GST-RBD4/CY in E. coli using an antibody against red opsin. (B) Western blot of purified red opsin from E. coli by itself and incubated with purified RBD4/CY, RBD4, and CY using an antibody against red opsin. Lanes 4 and 5 represent incubations with opsin purified from E. coli in the absence of detergent. (C) Western blot of red opsin purified from COS cells by itself and incubated with purified RBD4/CY. (D) Western blot of an aliquot of crude retinal extracts with an antibody against red opsin. A major 65-kDa opsin isoform was produced when red opsin was expressed either in E. coli or in COS cells. This opsin isoform may be converted to the 51-kDa RBD4/CY-binding-competent form by the concerted action of RBD4 and CY. The 65-kDa opsin isoform is a very minor species in retinal extracts, while the RBD4/CY-binding-competent form of opsin (51 kDa) is undetectable. In contrast, the 49.5-kDa (native) isoform is the major opsin isoform in retinal extracts. The 86-kDa and 34-kDa bands may represent SDS/PAGE-resistant opsin dimers and collapsed isoforms of red opsin, respectively. High molecular weight SDS/PAGE-resistant aggregates of opsin were seen in retinal extracts (D) and with opsin expressed in COS cells (C). All samples were run on the same SDS/polyacrylamide gel. CL, NT, BD, NN, and DM represent, respectively, the 34-kDa collapsed, 49.5-kDa native, 51-kDa RBD4/CY-binding-competent form, 65-kDa nonnative, and dimers of red opsin. R.E., bovine retinal extract.
The red opsin purified from E. coli was mainly the 65-kDa isoform (Fig. 4A, lane 1), whereas in bovine retinal extracts, the predominant red opsin species was the 49.5-kDa isoform (Fig. 4D) (38). The predicted molecular mass of human red opsin is 47.1 kDa. Western blot analysis of COS cell extracts expressing red opsin in the absence of RBD4/CY also revealed strong expression of the 65-kDa isoform (Fig. 4C, lane 1) but not of the 49.5-kDa isoform. We also observed a minor opsin isoform of 34 kDa in retinal extracts and extracts of expressing E. coli and COS cells. This isoform was not produced when we purified unfused red opsin with GST-RBD4/CY from E. coli coexpressing these proteins.
Finally, to confirm the previous results and establish that this RBD4/CY-induced modification of red opsin was not mediated by any additional eukaryotic cellular factors, we investigated the effects in vitro of purified RBD4/CY and fragments thereof on red opsin purified from both E. coli and COS cells. Addition of RBD4/CY to red opsin in vitro led to the production of a 51-kDa opsin isoform (Fig. 4 B and C). This effect was less pronounced with opsin purified from COS cells (Fig. 4C, lane 2). The RBD4/CY-induced modification of red opsin was not influenced by the addition of detergent during purification of opsin (Fig. 4B, lanes 4 and 5). The CY of RBD4/CY alone was insufficient to induce this modification.
DISCUSSION
The results presented here establish that RBD4-C is the binding domain for red opsin (Fig. 2). The N-RBD4 domain, in concert with CY, significantly enhances the binding of opsin to RBD4-C (Fig. 2). Thus, N-RBD4 and CY act as chaperones for red opsin to likely facilitate the interaction between RBD4-C and opsin. Removal of N-RBD4 from RBD4/CY abolishes its binding to opsin at 4°C. This binding can be significantly restored by the further removal of CY from RBD4-C/CY. This suggests that removal of N-RBD4 from RBD4/CY may alter the interaction of RBD4-C with CY. This change in interaction between RBD4-C and CY may sterically hinder the binding of RBD4-C to opsin in the RBD4-C/CY construct. The steric hindrance by CY on RBD4-C binding to opsin could be partially overcome by raising the incubation temperature (Fig. 2 Right), possibly due to partial “melting” of type I cyclophilin tertiary structure. In vivo, coexpression of type I cyclophilin (RBD4-C/CY) with red opsin leads to an increase in the production of functional visual pigment similar to that observed with RBD4/CY. Thus, the temperature-dependent binding and chaperone activity in vitro of RBD4-C/CY to opsin is consistent with the increased production in vivo of functional pigment at physiological temperature (Fig. 3). We also have shown previously that the PPIase activity of CY is reduced by about 40% in the presence of N-RBD4, likely due to a more restricted access of the short peptidyl prolyl-substrate to the active site of CY (33). Taken together, the data suggest that binding and interconversion of opsin by RBD4/CY may represent two coupled, but physically separate, processes involving opsin binding to RBD4 and CY-dependent modification of opsin.
It is possible that the chaperone activity of RBD4 extends also to Ran-GTPase. For example, the RanBP1 protein homologous to RBD4 also binds Ran in a GTP-dependent fashion (37). RanBP1 has been shown to stabilize the native conformation of Ran-GTP without affecting its intrinsic hydrolysis activity, likely by preventing the partial denaturation of Ran, and thus, the premature release of GTP from Ran (42). This suggests that RBD4 also may have a chaperone activity on Ran-bound nucleotide.
Attempted expression of unfused red opsin in E. coli cells resulted in the isolation of no recombinants, suggesting a possible cytotoxic effect of this protein. When plasmids encoding red opsin and RBD4/CY were used in a cotransformation experiment, cells expressing both proteins were readily isolated. This shows that RBD4/CY can rescue the apparent cytotoxic effect of unfused red opsin in E. coli, which supports the proposed role of RBD4/CY as a foldase and chaperone for red opsin. Expression of red opsin fused to GST by itself also was tolerated by E. coli cells. Expression of red opsin alone as a GST fusion, followed by cleavage of the GST fusion partner, resulted in an opsin isoform with an apparent molecular mass of 65 kDa. In contrast, coexpression of RBD4/CY with unfused red opsin in E. coli (Fig. 4A), or incubation of red opsin with RBD4/CY purified from E. coli (Fig. 4B) or COS cells (Fig. 4C) resulted in the production of a 51-kDa opsin isoform that is competent to bind RBD4/CY. The 65- and 51-kDa red opsin species were barely and not detectable in bovine retinal extracts, respectively (Fig. 4D). The major opsin isoform in retinal extracts is the 49.5-kDa form. Red opsin purified from COS cells was a poorer substrate than the opsin expressed in bacteria, possibly due to its extensive heterogeneous glycosylation as shown by the typical broader SDS/PAGE bands (Fig. 4C). The striking sharpness of the 51-kDa opsin band produced in the presence of RBD4/CY suggests that only minor unglycosylated and/or core-glycosylated red opsin produced in COS cells may be competent to be modified by RBD4/CY. As in retinal extracts (38), this conversion in vitro was enhanced by the presence of CY, but CY by itself did not produce a stable 51-kDa opsin isoform (Fig. 4B). Thus, the conversion of red opsin by RBD4/CY is a direct effect, with no requirements for additional cellular factors. In addition, the concerted action of RBD4 and CY is required to convert possibly different opsin isoform(s) in retinal extracts (38) and expressed in E. coli and COS cells to the 51-kDa form.
Still another isoform of human red opsin with an apparent molecular mass of 34 kDa was observed in different preparations of opsin from E. coli, COS cells, and retinal extracts (Fig. 4). This 34-kDa isoform may represent a compact (collapsed) conformer that excludes SDS detergent from its core. Copurification of red opsin expressed in E. coli with GST-RBD4/CY blocked the formation of this 34-kDa form. We also have previously shown that the 51-kDa RBD4/CY binding-competent isoform of bovine R/G opsin has no tendency to form a multimeric ladder on SDS/PAGE (e.g., compare Fig. 4 A and B with C and D) (38). We suggest that suppression of the formation of both the 34-kDa isoform and the ladder-pattern of red opsin self-aggregates, in addition to conversion of opsin to the 51-kDa isoform, represent independent physical conversions of opsin by RBD4/CY. We have proposed that this modification represents CY-PPIase mediated isomerization of one or more proline residues within the opsin molecule. This interpretation is supported by the observation that point mutations in the RBD4/CY PPIase catalytic-domain prevent conversion of bovine R/G opsin to the 51-kDa isoform (38), and that this shift requires CY (Fig. 4B) (38).
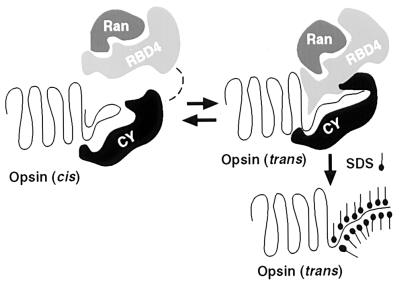
Both the 65-kDa and 49.5-kDa opsin isoforms may represent stable kinetically or thermodynamically trapped isoforms of opsin in slow conformational equilibrium with the 51-kDa form. Fig. 5 depicts a possible mechanism for this conformational trapping. In brief, RBD4 specifically binds the 51-kDa form of red opsin, possibly preventing its reisomerization to the 49.5- or 65-kDa forms. According to this model, CY influences the kinetics of this conversion by catalyzing cis–trans isomerization of proline residues in red opsin, while the RBD4 domain shifts the equilibrium toward the 51-kDa form by specifically binding to this isoform. We propose that addition of SDS detergent to the RBD4/CY-opsin complex stabilizes this 51-kDa isoform by denaturing the protein in this altered conformation, preventing subsequent spontaneous prolyl isomerization. The presence of two distinct electrophoretic isoforms of red opsin that may be converted into a third isoform suggests that at least two proline residues in red opsin may be undergoing isomerization.
Figure 5.
Schematic model of the interaction between the RBD4/CY supradomain and opsin. In the absence of added RBD4/CY, the majority of R/G opsin is in the cis-isomerized form. The prolyl-foldase, CY, induces cis to trans isomerization of one or more peptidyl-prolyl bonds in opsin, resulting in a form that is competent to bind RBD4. CY also is required as a chaperone to stabilize the interaction between opsin and RBD4. SDS stabilizes the trans form of opsin, which has a low propensity for self-aggregation. The cellular role of Ran-GTPase in opsin folding/processing is still unclear. It may be involved in the nuclear export of opsin mRNA, and with RBD4, may act as a CY-mediated coupling factor between transcription and translation of opsin(s) (38).
This model has implications beyond the processing of red opsin in cone cells. Recently, the free monomeric CY-A-binding loop domain of the N-terminal region of the HIV-1 capsid protein was shown to contain kinetically trapped prolyl conformations (43). In this case, CY-A is proposed to modulate the state of the prolyl “molecular switch” that may play a critical role during virion morphogenesis (43). It is possible that a similar mechanism operates in the formation of the RBD4/CY complex, where the concerted action of the CY/PPIase-mediated prolyl modification of red opsin and binding to RBD4 is critical to load and “lock” the opsin cargo onto RBD4. This system may serve as a model to understand other potential homologous molecular mechanisms and function of other proteins. For example, Pin1 (15) and its related proteins (44–46) contain PPIase and contiguous WW domains. The WW module of the Yes-associated protein (YAP) protein has been shown to bind proline-rich substrates (47). It has been suggested that the WW protein-binding module may confer specificity to the adjacent PPIase catalytic domain (15). The development of an accessible heterologous coexpression system for RBD4/CY and opsin described in this report may pave the way to understand the structural basis of the RBD4/CY-mediated modification of opsin. This is also important because many of the mutations that lead to retinal degeneration in a variety of species involve poorly understood defects in opsin biogenesis (29, 48–55). It is possible that some mutations causing retinal degeneration may affect, directly or indirectly, the interaction between opsin and RanBP2.
Acknowledgments
We gratefully acknowledge M. Kung for excellent technical assistance and J. Nathans for the generous gifts of hs7 cDNA clone and JH 492 antisera. We thank R. Molday for the monoclonal antibody 4D2. This work was funded by grants from the Fight for Sight, Inc., in memory of Silas Adelsheim and Wise Peggy, Research Division of Prevent Blindness America, and the National Eye Institute, and by an American Cancer Society institutional grant and the Dan Charitable Trust Fund for Research in Bilogical Sciences [Japan].
Footnotes
Abbreviations: R/G opsin, red/green opsin; GST, glutathione S-transferase; RanBP2, Ran-binding protein 2; RBD4, Ran-binding domain 4; CY, cyclophilin domain; PPIases, peptidyl-prolyl cis–trans isomerases; N-RBD4, N-terminal half of RBD4; RBD4-C, C-terminal half of RBD4.
References
- 1.Schmid F. Annu Rev Biophys Biomol Struct. 1993;22:123–143. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 2.Fischer G. Angew Chem Int Ed Engl. 1994;33:1415–1436. [Google Scholar]
- 3.Fischer G. Angew Chem. 1994;106:1479–1501. [Google Scholar]
- 4.Fischer G. Angew Chem Int Ed Engl. 1994;63:67–118. [Google Scholar]
- 5.Rudd K, Sophia H, Koonin E, Plunkett G, Lazar S, Rouviere P. Trends Biochem Sci. 1995;20:12–14. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 6.Rassaow J, Pfanner N. Curr Biol. 1996;6:115–118. doi: 10.1016/s0960-9822(02)00437-2. [DOI] [PubMed] [Google Scholar]
- 7.Brandts J, Halvorson H, Brennan M. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 8.Schmid F, Baldwin R. Proc Natl Acad Sci USA. 1978;75:4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid F, Grafl R, Wrba A, Beintema J. Proc Natl Acad Sci USA. 1986;83:872–876. doi: 10.1073/pnas.83.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freskgard P-O, Bergenhem N, Jonsson B-H, Svensson M, Carlsson U. Science. 1992;258:466–468. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 11.Lilie H, Lang K, Rudolph R, Buchner J. Protein Sci. 1993;2:1490–1496. doi: 10.1002/pro.5560020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinfert A, Collins C, Menard R, Anderson S. Biochemistry. 1994;33:1668–1673. doi: 10.1021/bi00173a008. [DOI] [PubMed] [Google Scholar]
- 13.Kern G, Kern D, Schmid F, Fisher G. FEBS Lett. 1994;348:145–148. doi: 10.1016/0014-5793(94)00591-5. [DOI] [PubMed] [Google Scholar]
- 14.Timerman A, Wiederrecht G, Marcy A, Fleischer S. J Biol Chem. 1995;270:2451–2459. doi: 10.1074/jbc.270.6.2451. [DOI] [PubMed] [Google Scholar]
- 15.Lu K, Hanes S, Hunter T. Nature (London) 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 16.Brillantes A-M, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella M, Jayaraman T, Landers M, Ehrlich B, Marks A. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 17.Cameron A, Steiner J, Roskams A, Ali S, Ronnett G, Snyder S. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Li B-Y, Danielson P, Shah P, Rockwell S, Lechleider R, Martin J, Manganaro T, Donahoe P. Cell. 1996;86:435–444. doi: 10.1016/s0092-8674(00)80116-6. [DOI] [PubMed] [Google Scholar]
- 19.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C, Sodroski J, Gottlinger H. Nature (London) 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 20.Helekar S, Char D, Neff S, Patrick J. Neuron. 1994;12:179–189. doi: 10.1016/0896-6273(94)90162-7. [DOI] [PubMed] [Google Scholar]
- 21.Shieh B-H, Stamnes M, Seavello S, Harris G, Zuker C S. Nature (London) 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- 22.Schneuwly S, Shortridge R, Larrivee D, Ono T, Ozaki M, Pak W. Proc Natl Acad Sci USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heitman J, Movva N, Hall M. New Biol. 1992;4:448–460. [PubMed] [Google Scholar]
- 24.Rahfeld J-U, Schierhorn A, Mann K, Fisher G. FEBS Lett. 1994;343:65–69. doi: 10.1016/0014-5793(94)80608-x. [DOI] [PubMed] [Google Scholar]
- 25.Rahfeld J-U, Rucknagel K, Schelbert B, Ludwig B, Hacker J, Mann K, Fisher G. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- 26.Stoller G, Rucknagel K, Nierhaus K, Schmid F, Fischer G, Rahfeld J. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh C, Zydowsky L, McKeon F. J Biol Chem. 1992;267:13115–13118. [PubMed] [Google Scholar]
- 28.Larrivee D, Conrad S, Stephenson R, Pak W. J Gen Physiol. 1981;78:521–545. doi: 10.1085/jgp.78.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephenson R S, O’Tousa J, Scavarda N, Randall L, Pak W. In: The Biology of Photoreception. Cosens D J, Vince-Price D, editors. Cambridge: Cambridge Univ. Press; 1983. pp. 447–501. [Google Scholar]
- 30.Stamnes A, Shieh B-H, Chuman L, Harris L, Zuker C. Cell. 1991;65:219–227. doi: 10.1016/0092-8674(91)90156-s. [DOI] [PubMed] [Google Scholar]
- 31.Baker E, Colley N, Zuker C. EMBO J. 1994;13:4886–4895. doi: 10.1002/j.1460-2075.1994.tb06816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira P, Pak W. In: Degenerative Diseases of the Retina. Anderson R, La Vail M, Hollyfield J, editors. New York: Plenum; 1995. pp. 263–274. [Google Scholar]
- 33.Ferreira P, Hom J, Pak W. J Biol Chem. 1995;270:23179–23188. doi: 10.1074/jbc.270.39.23179. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. Nature (London) 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Manutis M, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 36.Coutavas E, Ren M, Oppenheim J, D’Eustachio P, Rush M. Nature (London) 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 37.Drivas G, Shih A, Coutavas E, Rush M, D’Eustachio P. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira P, Nakayama T, Pak W, Travis G. Nature (London) 1996;383:637–640. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- 39.Nathans J, Thomas D, Hogness D. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 40.Guan K, Dixon J. Anal Biochem. 1990;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Macke J, Merbs S, Zack D, Klaunberg B, Bennett J, Gearhart J, Nathans J. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 42.Bischoff F, Krebber H, Smirnova E, Dong W, Ponstingl H. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gitti R, Lee B, Walker J, Summers M, Yoo S, Sundquist W. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 44.Hanes S, Shank P, Bostian K. Yeast. 1989;5:55–72. doi: 10.1002/yea.320050108. [DOI] [PubMed] [Google Scholar]
- 45.Hani J, Stumpf G, Domdey H. FEBS Lett. 1995;365:198–202. doi: 10.1016/0014-5793(95)00471-k. [DOI] [PubMed] [Google Scholar]
- 46.Maleska R, Hanes S, Hackett R, Couet H, Miklos G. Proc Natl Acad Sci USA. 1996;93:447–451. doi: 10.1073/pnas.93.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Sudol M. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard D, Bowman V, Ready D, Pak W. J Neurobiol. 1992;23:605–626. doi: 10.1002/neu.480230602. [DOI] [PubMed] [Google Scholar]
- 49.Kurada P, O’Tousa J. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 50.Colley N, Cassil J, Baker E, Zuker C. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung C-H, Schneider B, Agarwal N, Papermaster D, Nathans J. Proc Natl Acad Sci USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung C-H, Davenport C, Nathans J. J Biol Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- 53.Sung C-H, Makino C, Baylor D, Nathans J. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsson J, Gordon J, Pawlyk B, Roof D, Hayes A, Molday R, Mukai S, Cowley G, Berson E, Dryja T. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 55.Li T, Snyder W, Olsson J, Dryja T. Proc Natl Acad Sci USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]