Abstract
Ion selectivity is critical for the biological functions of voltage-dependent cation channels and is achieved by specific ion binding to a pore region called the selectivity filter. In voltage-gated K+, Na+ and Ca2+ channels, the selectivity filter is formed by a short polypeptide loop (called the H5 or P region) between the fifth and sixth transmembrane segments, donated by each of the four subunits or internal homologous domains forming the channel. While mutagenesis studies on voltage-gated K+ channels have begun to shed light on the structural organization of this pore region, little is known about the physical and chemical interactions that maintain the structural stability of the selectivity filter. Here we show that in an inwardly rectifying K+ (IRK) channel, IRK1, short range interactions of an ion pair in the H5 pore loop are crucial for pore structure and ion permeation. The two residues, a glutamate and an arginine, appear to form exposed salt bridges in the tetrameric channel. Alteration or disruption of such ion pair interactions dramatically alters ion selectivity and permeation. Since this ion pair is conserved in all IRK channels, it may constitute a general mechanism for maintaining the stability of the pore structure in this channel superfamily.
Ion channels are specialized membrane proteins that catalyze the downhill flow of ions across cell membranes. K+ channels selectively conduct K+ ions and regulate the electrical activity of all living cells (1). They consist of a large family of proteins that include voltage-gated K+ channels and inwardly rectifying K+ (IRK) channels. Voltage-gated K+ channel subunits have six transmembrane segments, with a pore-forming H5 or P region between the fifth and sixth helix (2). IRK channel subunits, on the other hand, have only two putative transmembrane segments with an H5 region sandwiched in between (3). Despite their distinct molecular architecture, both classes of K+ channels are tetrameric assemblies of homo- or hetromeric subunits (4–8) and are highly selective for K+ ions (1). The high K+ selectivity is achieved by specific interactions of K+ ions with the ion selectivity filter (1), which is formed by four H5 pore loops contributed from each of the four subunits (9). The structure and function of the H5 loop have been studied extensively in voltage-gated K+ channels. It has been shown that mutations of many residues in the H5 loop render the channel nonselective among monovalent cations (10–12). Many pore-lining residues in the region have been identified using the substituted cysteine accessibility method (13–16). More recently, using peptide toxins of known three-dimensional structure as an yardstick, the spatial localization of some residues has been mapped (16, 17–20). However, we know little about the physical and chemical interactions within the channel protein that maintains the stability of the pore structure.
Although IRK channels share little overall sequence homology with their voltage-gated K+ channel relatives, many residues are conserved in the H5 region in both families, including a Gly-Tyr-Gly motif. This conservation suggests that the H5 loop is important for ion selectivity and permeation in IRK channels as well. Indeed, mutation of a conserved glycine residue in a G protein-gated IRK channel results in loss of K+ selectivity (21–23). However, compared with our knowledge on voltage-gated K+ channels, far less is known about the structure and function of the H5 loop in IRK channels. This lack occurs partly due to the fact that mutation of many H5 loop residues produce nonfunctional channels, as each mutation is repeated 4 times in the expressed tetrameric channel. To overcome this problem, we have employed a rescuing strategy by linking just one or two mutant subunits with wild-type subunits into tandem tetramers. Since most of the channels generated by this method are formed by the intramolecular assembly of the four subunits within the tetramer (5, 7) and thus contain only one or two mutant subunits, they are more likely to be functional and can thus be studied. In this work, we have used this rescuing strategy to study the functions of two oppositely charged residues in the H5 loop in a strongly rectifying channel IRK1 (24). We demonstrate that these two residues form an interacting ion pair critical for maintaining the stability of the pore structure and for ion selectivity and permeation.
MATERIALS AND METHODS
Construction of Point Mutants and Tandem Tetramers.
All site-directed mutations were produced by the oligonucleotide insertion method (25) and confirmed by sequencing. Tandem tetramers were constructed in three steps as described previously (7). First, four monomeric IRK1 cDNAs with unique restriction site(s) and/or linkers were constructed in pBluescript SK− (Strategene). In subunit A, a linker containing 10 glutamines plus the first 10 amino acids from IRK1 with a built-in PinAI site was ligated into the AflII–HindIII sites. In subunit B, a linker containing 10 glutamines and residues Thr-Arg bearing a MluI site was subcloned as described above, and a PinAI site was engineered at the beginning of the N terminus at the Asn-Arg residues. Subunit C was derived from subunit A but with a MluI site created immediately before the starting codon. Subunit D was derived from subunit B but without the linker. Second, the PinAI–HindIII fragment from subunit B was ligated in-frame into subunit A to create dimer A·B, and that from subunit D into subunit C to create dimer C·D. Third, the PstI–MluI fragment from A·B was subcloned into C·D to produce a tetrameric cDNA.
Oocyte Expression.
To obtain high level expression in oocytes, all constructs were subcloned from pBluscript SK− (the BamHI–HindIII fragment containing the entire coding region) into pGEMHE (5), which contains 5′ and 3′ untranslated regions of a Xenopus β-globin gene. cRNAs for all constructs were transcribed in vitro using the T7 RNA polymerase following linearization of the cDNAs with NheI. Oocytes isolated from Xenopus laevis were injected with 0.2–5 ng of cRNA, maintained in an 18°C incubator, and used for recordings 2–15 days after injection.
Electrophysiology.
Two electrode voltage-clamp recordings were performed by using the CA-1a oocyte clamp amplifier (Dagan Instruments, Minneapolis). Recording electrodes were filled with 3 M KCl. External solution contained 90 mM KCl, 2 mM MgCl2, and 10 mM Hepes (pH 7.3). When necessary, KCl was replaced with NaCl or N-methyl-d-glucamine (NMDG). Current records obtained in the NMDG solution were used for leak subtraction. In pH titration experiments, the desired pH value was obtained by the addition of KOH or HCl to the external solution and was measured with a Corning digital pH meter. Whole-cell current was first recorded in the control solution (pH 7.0) and then in the test solution (for 5–10 min, depending on pH) and back in control solution. Current was measured at the end of a 200-ms voltage step to −100 mV from a holding potential of −60 mV and was normalized by that obtained at pH 7. Each type of experiment was performed in 2–4 different batches of oocytes.
Single-channel recordings were performed using the Axopatch 200A amplifier (Axon Instruments, Foster City, CA). Oocyte was devitellinized and bathed in a solution containing 110 mM KCl, 10 mM Hepes, 9 mM EGTA, 1 mM EDTA, and 200 μM CaCl2; pH was adjusted to 7.3 with KOH (total K+ ≈140 mM). Recording glass pipettes pulled from pyrex glass tubes (Corning) were filled with a solution of 130 mM KCl, 10 mM Hepes, and 3 mM MgCl2; pH was adjusted to 7.3 with KOH (total K+ ≈140 mM), and had resistances of 6–10 MΩ. Currents were filtered at 1 KHz and digitized at 2.5 KHz. All experiments were performed at room temperature (22–25°C).
Data Analysis.
For the wild-type and WT·E138Q· E138Q·WT tetramer, pH titration data were fitted according to Hill equation I/ICon = Kdn/(Cn + Kdn), where C is the proton concentration, Kd is the equilibrium constant, and n is the Hill coefficient. For the WT·R148H·R148H·WT tetramer, data in the basic pH range (pH 7–10) were fitted to the equation I/ICon = 1 − 0.21 × Kdn/(Cn + Kdn), and those in the acidic pH range (pH 1–7) were fitted to the sum of two Hill equations, each with a fixed Hill coefficient of 2 according to I/ICon = A × Kd12/(C2 + Kd12) + B × Kd22/(C2 + Kd22), where A and B are the fractions of each component (A + B = 1). Data from different batches of oocytes were pooled together and are presented as mean ± SEM (number of oocytes).
RESULTS AND DISCUSSION
Functional Interactions Between a Pore Loop Ion Pair.
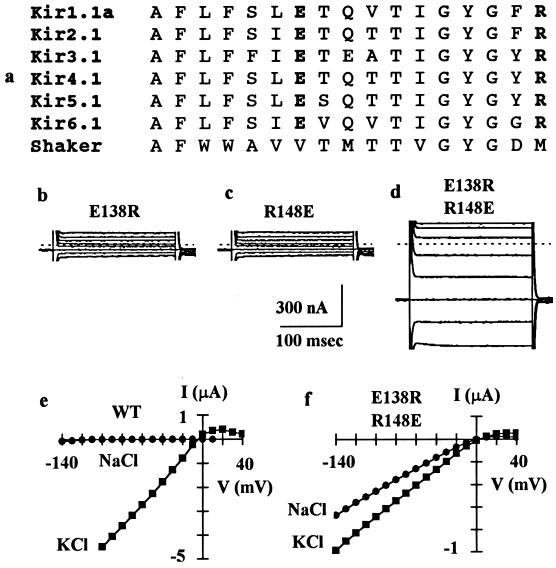
We focused our study on two charged residues in the H5 region, a negatively charged glutamate (Glu-138 in IRK1) and a positively charged arginine (Arg-148 in IRK1), since they are unique to and are conserved in all IRK channels cloned so far (Fig. 1a). Substitution of these two residues individually with many others produced nonfunctional channels in Xenopus oocytes, including mutations of Glu-138 to Asp, Gln, Cys, or Arg or of Arg-148 to Lys, His, Cys, or Glu (Fig. 1 b and c). This result may be due to one of the two reasons: (i) the mutations disrupt channel protein folding, assembly, or targeting such that the mutant subunit proteins never get to the plasma membrane; or (ii) the mutant subunits do form channels in the surface membrane, but the mutations cause the channels to become nonconducting. The latter appears to be the cause as Western blot analysis indicates that E138R and R148E subunits (the two mutants we tested) are expressed in the oocyte plasma membrane (data not shown).
Figure 1.
Functional interactions between a pair of pore loop residues. (a) Alignment of the H5 pore loop amino acid sequences from members of six subfamilies of inwardly rectifying K+ channels. See Doupnik et al. (3) for nomenclature. The IRK1 channel used in this study corresponds to Kir2.1. The amino acid sequence corresponds to number 132–148. The H5 region sequence of the voltage-gated Shaker K+ channel is also shown for comparison. (b–d) Whole-cell currents recorded by two-electrode voltage-clamp from oocytes injected with cRNA for E138R (b), R148E (c), or E138R/R148E (d). Currents were elicited by 200-ms voltage steps to −100 to +40 mV in 20-mV increments from a holding potential of −60 mV and are not leak-subtracted. The dashed line indicates zero current level. The current amplitude at −100 mV was 111 ± 7 nA (mean ± SEM, n = 11) for E138R, 119 ± 7 nA (n = 11) for R148E, and 671 ± 27 nA (n = 20) for E138R/R148E. Oocytes injected with cRNAs for E138D, E138Q, E138C, R148K, R148H, or R148C, like oocytes injected with cRNAs for E138R or R148E, produced currents not significantly different from uninjected oocytes, which had a basal current of 116 ± 21 nA (n = 21) at −100 mV. (e–f) Current–voltage relations in 90 mM external KCl or NaCl for the wild-type IRK1 channel or mutant E138R/R148E channel. Current was measured at the end of a 200-ms test pulse and was leak-subtracted. For the wild-type channel, the reversal potential was −5.9 ± 0.3 mV (mean ± SEM, n = 9) in KCl and −103 ± 3 mV (n = 9) in NaCl. For the mutant channel, the reversal potential was −4.8 ± 1.0 mV (n = 11) in KCl and −7.2 ± 1.2 mV (n = 11) in NaCl.
Since residues Glu-138 and Arg-148 carry opposite charges, we hypothesized that they might form an interacting ion pair. To test this possibility, we constructed a reverse double mutant by swapping the residues at the two positions. Interestingly, the double mutation restored channel activity, which still exhibited inward rectification (Fig. 1d). However, the E138R/R148E mutant channel differed from the wild-type channel in at least two ways. First, the whole-cell current amplitude was 50–100 times smaller than that of oocytes injected with similar amount of wild-type cRNA. Indeed, attempts to record the single-channel current of the mutant channel failed repeatedly, presumably due to a much reduced single-channel conductance; the wild-type channel has a conductance of ≈21 pS (24). Second, the mutant channel lost its K+ selectivity. Wild-type IRK1 channel was highly selective for K+ over Na+ and conducted little inward Na+ current (Fig. 1e), with a permeability ratio PNa/PK of 0.02 ± 0.005 (mean ± SEM, n = 9). By contrast, the mutant channel became permeable to Na+ with a PNa/PK of 0.91 ± 0.06 (n = 11) and conducted significant inward Na+ current (Fig. 1f). Despite these differences, the finding that the reverse double mutation was capable of restoring channel activity strongly suggests that residues Glu-138 and Arg-148 interact with each other and that such interaction is important for channel activity, possibly by contributing to the stability of the pore structure.
pH Titration Reveals Salt Bridge Formation Between Glu-138 and Arg-148.
One possible interaction between Glu-138 and Arg-148 is the formation of a salt bridge, which involves both short range electrostatic attraction and hydrogen bonding (Fig. 2a). We examined this possibility by testing the effect of external pH on current produced by the wild-type IRK1 channel. We reasoned that if Glu-138 and Arg-148 are involved in a salt bridge that is critical for maintaining the structural stability of the pore region, protonation of Glu-138 or deprotonation of Arg-148 should disrupt the salt bridge and hence destabilize the pore structure, thereby attenuating channel activity. Because the intrinsic pKa of arginine is ≈12, it is technically difficult to titrate its guanido group. On the other hand, glutamate has an intrinsic pKa of ≈4.3 and should be relatively easier to titrate, therefore, we focused our study on Glu-138. In general, an acidic residue involved in a stabilizing salt bridge will have a lower pKa than its intrinsic value (26). For example, the pKa of Asp-70, which forms a salt bridge with His-31 in T4 lysozyme, is lowered by ≈3.5 units relative to its intrinsic value of 3.9 (27).
Figure 2.
pH titration suggests formation of Glu-138–Arg-148 salt bridges. (a) The carboxyl group of Glu-138 and the guanido group of Arg-148 are proposed to form a salt bridge, which involves both short range electrostatic attraction and hydrogen bonding. (b) Effect of external pH on whole-cell current produced by the wild-type IRK1 channel, recorded by two-electrode voltage-clamp. Each data point is an average of 5–8 measurements. Solid line represents a least-squares fit to the Hill equation with a pKa of 1.97 and a Hill coefficient of 4.1.
Fig. 2b shows the effect of changing external pH on the whole-cell current of wild-type IRK1 channel expressed in oocytes. Channel activity was stable between pH 10 and 2.25, but declined sharply upon further lowering of pH. This attenuation was unlikely due to block of the channel by external protons, since it developed over 3–8 min and was not readily reversible (data not shown). The least-squares fit to the pH titration data yielded a pKa of 1.97 and a Hill coefficient of 4.1. The acidic shift of pKa is consistent with the protonation of an acidic residue involved in salt bridging, and a Hill coefficient of 4 suggests a highly cooperative effect of pH and is consistent with the tetrahomomeric composition of the IRK1 channel (7, 8).
To ascertain that the observed pH effect arose from protonation of Glu-138 and subsequent disruption of the Glu-138-Arg-148 ion pair interaction, we examined the effect on pH dependence of mutation of Glu-138 to glutamine or of Arg-148 to histidine. We took advantage of the fact that histidine has an intrinsic pKa of ≈7 and should be titratable in the pH range in our experiments. Since neither mutation produced functional homomeric channels (see Fig. 1 legend), they were functionally rescued using the tandem tetramer approach. Two tandem tetramers were constructed, one contained two subunits bearing the E138Q mutation (WT·E138Q·E138Q·WT), and the other contained two subunits bearing the R148H mutation (WT·R148H·R148H·WT). Channels generated by both tetramers exhibited altered pH dependence compared with the wild-type (Fig. 3). Neutralizing two of the four Glu-138 residues to glutamine in the WT·E138Q·E138Q·WT channel did not perturb the pKa of pH titration (pKa = 1.9) but reduced the Hill coefficient from 4 (in the wild-type) to 2, coinciding with the presence of two remaining intact Glu-138–Arg-148 ion pairs in the tetrameric channel.
Figure 3.
Mutation of Glu-138 or Arg-148 alters pH titration. (a) Effect of external pH on whole-cell current recorded by two-electrode voltage-clamp from oocytes expressing the WT·E138Q·E138Q·WT or WT·R148H·R148H·WT tetramer. Data for the wild-type channel are also shown for comparison. Each data point is an average of 5–8 measurements. Solid lines represent least-squares fit of the data for each channel type (see text for detail). (b) A close-up of the response in the acidic pH range.
A more complex pH effect was observed for channels formed by the WT·R148H·R148H·WT tetramer, in which two Arg-148 residues were changed to histidine (Fig. 3). In the acidic pH range, the titration data appeared to have two components and could be fitted by the sum of two Hill functions, each with a fixed Hill coefficient of 2. One component (27%) has a pKa of 3.3, presumably resulting from protonation of the two Glu-138 residues in the Glu-138–His-148 ion pair. The smaller acidic shift of pKa from its intrinsic value is expected, because Glu-138 is expected to have a weaker interaction with a histidine than an arginine. The other component (73%) has a pKa of 2.1, similar to that for the wild-type channel, presumably resulting from protonation of Glu-138 residues in the two remaining Glu-138-Arg-148 ion pairs.
In contrast to the wild-type channel or the WT· E138Q·E138Q·WT channel, current generated by the WT·R148H·R148H·WT channel was decreased by 21% (±3, n = 5) when external pH was increased from 7 to 10. A least-squares fit to the titration data with a Hill equation yielded a Hill coefficient of 1.7 and a pKa of 8.5. Such an elevated pKa is expected for a histidine residue involved in a stabilizing salt bridge. The decrease of current is thus most likely a result of deprotonation of the two substituting histidine residues and subsequent alteration of the Glu-138–His-148 ion pair interaction. The maximum magnitude of current reduction is close the first component (27%) of inhibition induced by pH acidification, indicating similar impact of protonation of Glu-138 or deprotonation of His-148 in the Glu-138–His148 ion pair. Regardless of the mechanistic interpretation, the observation that mutation of either Glu-138 or Arg-148 alters the pH dependence provides a strong support for the notion of salt bridge formation between the two residues.
Disruption of Glu-138–Arg-148 Salt Bridge Alters Ion Permeation.
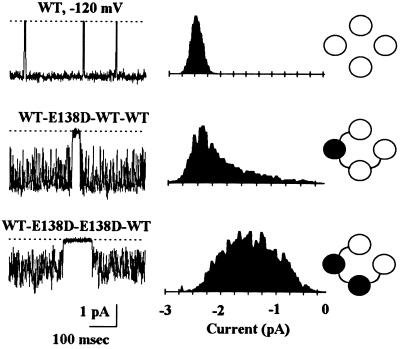
The mutagenesis and pH titration studies presented above indicate that the Glu-138–Arg-148 salt bridge is crucial for the IRK1 channel activity. Indeed, alteration of just one or two such ion pair interactions can produce dramatic consequences on ion permeation. Fig. 4 demonstrates the effect on single-channel current of substituting Glu-138 with aspartate, a conservative mutation that retains the negative charge but shortens the side chain by ≈1.5 Å, which is enough to disrupt hydrogen bonding. Changing just one of the four Glu-138 residues to aspartate induced rapid flickering of the single open channel; mutating two to aspartate caused further flickering and almost halved the mean current amplitude (Fig. 4); changing all four glutamates to aspartate resulted in nonfunctional channels (see Fig. 1 legend). The rapid flickering in the mutant channels was not caused by block of channel pore from either side by ions other than K+, since it persisted in inside-out patches exposed to isotonic KCl solutions on both sides (data not shown). Instead, it most likely reflects alteration of K+ binding to the pore as a result of changes in the pore structure, induced by weakened ion pair interactions. However, despite dramatic changes in single-channel property, channels formed by the WT·E138D·E138D·WT tetramer remained highly selective for K+ ions and exhibited similar sensitivities to internal Mg2+ and polyamines as the wild-type channel (data not shown), indicating that any structural change caused by the E138D mutation is local and subtle.
Figure 4.
Disruption of one or two Glu-138–Arg-148 ion pairs alters ion permeation. (Left) Cell-attached single-channel currents recorded at −120 mV from oocytes expressing the wild-type channel or mutant channels generated by the WT·E138D·WT·WT or WT·E138D· E138D·WT tetramer. (Center) The corresponding all point amplitude histogram. The presumed channel type is depicted on the Right.
Electrostatic and hydrogen bonding interactions between ionized amino acids are known to be important for enzyme catalysis, subunit interaction, and protein folding (28). In this study, we demonstrate that a Glu–Arg ion pair in the H5 pore loop is critical for the proper functioning of IRK1 channel, presumably by forming salt bridges that contribute to the structural stability of the pore region formed by the H5 pore loops (i.e., the ion selectivity filter). The pKa shift of Glu-138 from an intrinsic value of 4.3 to 2 indicates that a single Glu-138–Arg-148 salt bridge contributes ≈3.1 kcal/mol (given by ΔΔGtitration = −2.303RTΔpKa). Since there are four Glu-138–Arg-148 ion pairs in the tetrameric channel, their total contribution to the structural stability of the pore is substantial. Disruption of these ion pair interactions, either by site-directed mutation or by changes of external pH, may lead to alteration of the local pore structure and, hence, a reduction or total loss of channel activity. The susceptibility of the Glu-138–Arg-148 ion pair to external protons suggests that it is exposed to the high dielectric aqueous solution. This may help explain its vulnerability to mutations, since even conservative mutations such as glutamate to aspartate or arginine to lysine will greatly decrease the electrostatic and hydrogen bonding interactions in such an exposed ion pair. Exposure to solution also provides an opportunity for the side chains to interact with permeant K+ ions. The conservation of this ion pair in all IRK channels may result from an evolution pressure; unlike voltage-gated Na+, Ca2+, and K+ channels, IRK channel subunits are smaller and possess only two putative transmembrane segments. By utilizing the stabilizing energy of the pore loop ion pairs, IRK channels may adopt a common mechanism to maintain the stability of the tertiary structure of the ion selectivity filter.
Acknowledgments
We thank colleagues in our laboratory for helpful discussions and comments. This work was supported by fellowships from the Howard Hughes Medical Institute and National Institutes of Health (to J.Y.) and a grant from the National Institutes of Health. Y.N.J. and L.Y.J. are Howard Hughes Medical Institute investigators.
Footnotes
Abbreviation: IRK channel, inwardly rectifying K+ channel.
References
- 1.Hille B. Ionic Channels of Excitable Membranes. 2nd Ed. Sutherland, MA: Sinauer; 1992. p. 607. [Google Scholar]
- 2.Jan L Y, Jan Y N. Cell. 1992;69:715–718. doi: 10.1016/0092-8674(92)90280-p. [DOI] [PubMed] [Google Scholar]
- 3.Doupnik C A, Davidson N, Lester H A. Curr Opin Neurobiol. 1995;5:268–277. doi: 10.1016/0959-4388(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 4.MacKinnon R. Nature (London) 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- 5.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 6.Li M, Unwin N, Stauffer K A, Jan L Y, Jan Y N. Curr Biol. 1993;4:110–115. doi: 10.1016/s0960-9822(94)00026-6. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Jan Y N, Jan L Y. Neuron. 1995;15:1441–1447. doi: 10.1016/0896-6273(95)90021-7. [DOI] [PubMed] [Google Scholar]
- 8.Glowatzki E, Fakler G, Brandel U, Rexhausen U, Zenner H P, Ruppersberg J P, Fakler B. Proc R Soc London Ser B. 1995;261:251–261. doi: 10.1098/rspb.1995.0145. [DOI] [PubMed] [Google Scholar]
- 9.MacKinnon R. Neuron. 1995;14:889–892. doi: 10.1016/0896-6273(95)90327-5. [DOI] [PubMed] [Google Scholar]
- 10.Yool A J, Schwartz T L. Nature (London) 1991;349:700–704. doi: 10.1038/349700a0. [DOI] [PubMed] [Google Scholar]
- 11.Taglialatela M, Drewe J A, Kirsch G E, DeBiasi M, Hartmann H A, Brown A M. Pflügers Arch. 1993;423:104–112. doi: 10.1007/BF00374967. [DOI] [PubMed] [Google Scholar]
- 12.Heginbotham L, Lu Z, Abramson T, MacKinnon R. Biophys J. 1994;66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kürz L L, Zuhlke R D, Zhang H J, Joho R H. Biophys J. 1995;68:900–905. doi: 10.1016/S0006-3495(95)80266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lü Q, Miller C. Science. 1995;268:304–307. doi: 10.1126/science.7716526. [DOI] [PubMed] [Google Scholar]
- 15.Pascual J M, Shieh C C, Kirsch G E, Brown A M. Neuron. 1995;14:1055–1063. doi: 10.1016/0896-6273(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 16.Gross A, MacKinnon R. Neuron. 1996;16:399–406. doi: 10.1016/s0896-6273(00)80057-4. [DOI] [PubMed] [Google Scholar]
- 17.Aiyar J, Withka J M, Rizzi J P, Singleton D H, Andrews G C, Lin W, Boyd J, Hanson D, Simon M, Dethlefs B, Lee C L, Hall J E, Gutman G A, Chandy G. Neuron. 1995;15:1169–1181. doi: 10.1016/0896-6273(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 18.Hidalgo P, MacKinnon R. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 19.Naranjo D, Miller C. Neuron. 1996;16:123–130. doi: 10.1016/s0896-6273(00)80029-x. [DOI] [PubMed] [Google Scholar]
- 20.Ranganathan R, Lewis J H, MacKinnon R. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 21.Slesinger P, Patil N, Liao Y J, Jan Y N, Jan L Y, Cox D R. Neuron. 1996;16:321–331. doi: 10.1016/s0896-6273(00)80050-1. [DOI] [PubMed] [Google Scholar]
- 22.Kofuji P, Hofer M, Millen K J, Millonig J H, Davidson N, Lester H A, Hatten M. Neuron. 1996;16:941–952. doi: 10.1016/s0896-6273(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 23.Navarro B, Kennedy M E, Velimirovic B, Bhat D, Peterson A S, Clapham D E. Science. 1996;272:1950–1953. doi: 10.1126/science.272.5270.1950. [DOI] [PubMed] [Google Scholar]
- 24.Kubo Y, Baldwin T J, Jan Y N, Jan L Y. Nature (London) 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- 25.Richards J H. In: Direct Mutagenesis: A Practical Approach. McPherson M J, editor. Oxford: Oxford Univ. Press; 1991. pp. 199–215. [Google Scholar]
- 26.Yang A-S, Honig B. Curr Opin Struct Biol. 1992;2:40–45. [Google Scholar]
- 27.Anderson D E, Becktel W J, Dahlquist F W. Biochemistry. 1990;29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 28.Perutz M F. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]