Abstract
The function of acyl carrier protein (ACP) in mitochondria isolated from pea leaves has been investigated. When pea leaf mitochondria were labeled with [2-14C]malonic acid in vitro, radioactivity was incorporated into fatty acids, and, simultaneously, ACP was acylated. [1-14C]Acetate was much less effective as a precursor for fatty acid synthesis, suggesting that mitochondria do not possess acetyl-CoA carboxylase. The incorporation of radioactivity from [2-14C]malonate into fatty acids and the labeling of ACP were inhibited by cerulenin and required ATP and Mg2+. These findings indicate that plant mitochondria contain not only ACP, but all enzymes required for de novo fatty acid synthesis. Over 30% of the radioactive products from pea mitochondria labeled with [2-14C]malonate were recovered in H protein, which is a subunit of glycine decarboxylase and contains lipoic acid as an essential constituent. In similar experiments, the H protein of Neurospora mitochondria was also labeled by [2-14C]malonate. The labeling of pea H protein was inhibited by addition of cerulenin into the assay medium. Together, these findings indicate that ACP is involved in the de novo synthesis of fatty acids in plant mitochondria and that a major function of this pathway is production of lipoic acid precursors.
Keywords: acyl carrier protein, H protein, glycine decarboxylase, Neurospora, pea
Acyl carrier proteins (ACPs) have been reported to occur in mitochondria of Neurospora (1, 2), Saccharomyces cerevisiae (3), plants (4, 5), and bovine heart muscle (6). The function of these proteins in mitochondria has been an enigma. ACPs are best known as protein cofactors that are essential for de novo fatty acid synthesis in all organisms (7–10). ACPs carry acyl chains through the condensation, reduction, and dehydration steps of the fatty acid synthesis pathway and are also involved in desaturase, hydrolase, and acyltransferase reactions. In addition to these well known roles in fatty acid metabolism, ACPs also function in syntheses of membrane-derived oligosaccharides (11) and polyketides (12) and in the activation of hemolysin, an Escherichia coli membrane-targeted toxin (13). Furthermore, in mitochondria of Neurospora (2) and bovine heart muscle (6), ACP has been identified as a component of the NADH:ubiquinone oxidoreductase complex (complex I), and the acylated form of mitochondrial ACP (mtACP) is found to be associated with a peripheral part of complex I (14). Recently, Schneider et al. (10) disrupted the genes encoding the mtACPs of S. cerevisiae and Neurospora. In S. cerevisiae, which does not contain complex I and in which mtACP is localized in the mitochondrial matrix, the loss of mtACP leads to a pleiotropic respiratory-deficient phenotype, whereas in Neurospora, the disruption of the gene for mtACP results in a respiratory-defective phenotype and disturbed assembly of complex I.
Based on incorporation of [14C]malonate into fatty acids and acyl-mtACP, it has also been reported that mtACP is involved in de novo fatty acid synthesis in mitochondria of Neurospora (15). Furthermore, Shintani and Ohlrogge (5) found that mtACP from Arabidopsis was able to function as a cofactor for the chloroplast fatty acid synthase in vitro. However, it has not been demonstrated that mtACP is involved in de novo fatty acid synthesis in mitochondria of plant cells nor has the function of this pathway been elucidated in any organism. Because most or all mitochondrial membrane fatty acids are believed to originate outside the mitochondria, the role of the mitochondrial fatty acid biosynthetic pathway is unknown.
In work reported here, we examined fatty acid synthesis in pea mitochondria and investigated the function of mtACP. Our results indicated that a complete pathway for de novo fatty acid synthesis occurs in pea mitochondria and that mtACP is involved in the process. Furthermore, our data suggest that a major part of the de novo synthesized fatty acids is used for biosynthesis of lipoic acid, an essential cofactor of mitochondrial enzymes such as glycine decarboxylase and pyruvate dehydrogenase. These results establish a function for biosynthesis of fatty acids in mitochondria and begin to define the subcellular organization of lipoic acid biosynthesis in eukaryotes.
MATERIALS AND METHODS
Isolation of Mitochondria.
Pea (Pisum sativum L. cv. Little Marvel; Burpee Seeds, Waminster, PA) seedlings were grown at 20°C under a 8 h light/16 h dark cycle. Mitochondria were isolated from ≈400 g of 14-day-old green seedlings as described previously (5). Contamination of mitochondrial preparations by chloroplast proteins was estimated by chlorophyll content in the preparations as measured by the method of Arnon (16). Contamination was calculated based on the protein content of the mitochondrial preparation and a ratio of 7 mg of chloroplast protein per mg of chlorophyll.
Mitochondria of Neurospora crassa were prepared from exponentially growing hyphae of the standard 74-OR23-1A laboratory strain by the flotation gradient method of Lizardi and Luck (17), as described by Lambowitz (18) and modified by Infanger and Bertrand (19).
Labeling of Mitochondria.
Mitochondria corresponding to 400 μg mitochondrial protein were mixed with various components to give the following final concentrations in a total volume of 200 μl: 50 mM TES-KOH (pH 7.2), 10 mM MgCl2, 0.3 M sorbitol, 0.1% (wt/vol) BSA, 1 mM each ATP, ADP, NADH, NADPH, CoA, and DTT, and 1 μCi (1 Ci = 37 GBq) of [2-14C]malonic acid (specific activity, 55 mCi/mmol) or 1 μCi of [1-14C]acetate (specific activity, 47 mCi/mmol). The reaction mixture was incubated for 1 h at room temperature (≈23°C). After the incubation, mitochondria were collected by centrifugation at 15,000 × g for 5 min. The mitochondrial pellets were washed by resuspension with 500 μl of 50 mM TES-KOH (pH 7.2) and 0.3 M sorbitol, and by centrifugation at 15,000 × g for 5 min. The pelleted mitochondria were used for further analysis. When matrix and membrane fractions were prepared, mitochondria were disrupted by sonication and centrifuged at 100,000 × g for 60 min. The resultant supernatant was used for further analysis as a matrix fraction. The pellet was washed in 50 mM TES-KOH (pH 7.2) and 0.3 M sorbitol, and used as a membrane fraction.
Protein concentrations were determined by the method of Bradford (20) using BSA as standard.
Analysis of 14C-Labeled Proteins.
The mitochondria labeled with [2-14C]malonic acid or [1-14C]acetic acid were suspended in 80 μl of 1× SDS/PAGE sample buffer and boiled for 3 min. An aliquot of 40 μl was subjected to electrophoresis on 15% SDS/polyacrylamide gels and blotted onto nitrocellulose membrane filters as described by Battey and Ohlrogge (21). Radioactivity on the blots was detected and analyzed by PhosphorImager (Molecular Dynamics). 14C-Labeled acyl-mtACPs were identified by comparing migration to 14C-labeled 16:0-mtACPs standard synthesized in vitro using E. coli acyl-ACP synthetase (22). The 14C-labeled acyl groups bound to mtACPs were analyzed as follows. The bands containing the labeled mtACPs were cut from the nitrocellulose filters. The acyl-mtACPs were derivatized to fatty acid methyl esters (FAMEs) by dissolving the nitrocellulose in 1.5 ml of 0.5 M sodium methoxide in methanol and incubated at room temperature for 1 h. The derivatization was stopped by adding 1.5 ml of 125 mM H2SO4, and the FAMEs were then extracted three times with 3 ml of hexane. Individual species of FAMEs derived from acyl-mtACPs were separated and analyzed on TLC plates as described below.
Western blot analysis was carried out according to Post-Beittenmiller et al. (23). Immunoprecipitation of H protein of glycine decarboxylase was performed as described by Roesler et al. (24).
Fatty Acid Analysis.
Mitochondria labeled with [2-14C]malonic acid or [1-14C]acetic acid were suspended in 200 μl of H2O and saponified with 0.1 volume of 10 M KOH for 15 min at 65°C, following which 0.2 volume of 5 M H2SO4 and 1 volume of a solution of 1 mM palmitic acid in 1 M acetic acid in isopropanol were added to the sample. The saponified fatty acids were extracted three times with 2 volumes of hexane and analyzed by scintillation counting. The hexane was evaporated in a N2 stream, and fatty acids were derivatized to FAMEs by 1 ml of boron trichloride in methanol (Sigma) at 90°C for 10 min. The obtained FAMEs were extracted three times with 2 ml of hexane after addition of 1 ml of H2O. The FAMEs were separated on KC18 reversed-phase TLC plates (Whatman) using as solvent system, acetonitrile:methanol:water (65:35:0.5, vol/vol) and analyzed by PhosphorImager. Individual FAMEs were identified by comparing their migration to those of authentic FAMEs. To minimize loss of short chain FAME, all evaporations were at room temperature or lower temperatures and were stopped immediately upon removal of solvent.
RESULTS
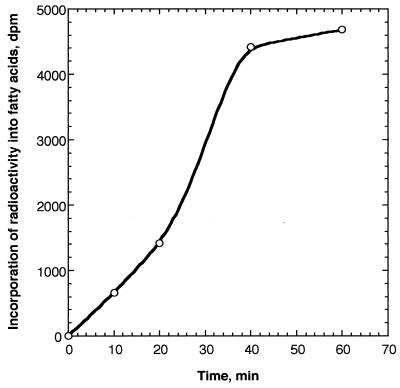
To investigate whether fatty acid synthesis takes place in plant mitochondria, we purified mitochondria from pea leaves and incubated them in vitro with [2-14C]malonic acid or [1-14C]acetate. Fig. 1 shows the time course of incorporation of radioactivity from [14C]malonic acid into fatty acids. The incorporation of radioactivity into fatty acids increased approximately linearly with incubation times up to 40 min. After 1 h of incubation, 4700 dpm radioactivity, which corresponds to 0.2% of added [14C]malonate, was incorporated into saponifiable fatty acids. When the mitochondria were labeled with [14C]acetate, radioactivity was not significantly incorporated into fatty acids (data not shown), suggesting that the pea mitochondria do not possess acetyl-CoA carboxylase. This finding is consistent with the results of Baldet et al. (25) that acetyl-CoA carboxylase activity was not detected in pea mitochondria. Although the rate of precursor incorporation into fatty acids by mitochondria (≈0.2 nmol·h−1·mg of protein−1) is much less than that in chloroplasts (26), these data demonstrate that fatty acid synthesis does occur in pea mitochondria. The synthesis of fatty acids is not due to contamination of mitochondria by chloroplasts because: (i) by chlorophyll analysis, mitochondrial preparations were found to contain <0.8% chloroplast protein; (ii) chloroplasts are highly active in acetate incorporation into fatty acids, but very inactive with malonate in contrast to the opposite pattern we observed; and (iii) chloroplasts produce mainly 16:0, 18:0, and 18:1, in contrast to the shorter chain products we observed. Furthermore, because malonyl-CoA, which does not cross membranes, was much less effective than malonate as a precursor (data not shown), the activity we observed is not attributable to contamination of the mitochondria by fatty acid synthase enzymes released from broken chloroplasts or other sources.
Figure 1.
Time course of incorporation of [2-14C]malonate into fatty acids. Pea mitochondria (400 μg of protein) were labeled with [2-14C]malonic acid for the designated times, and radioactivity incorporated into fatty acids was determined by scintillation counting.
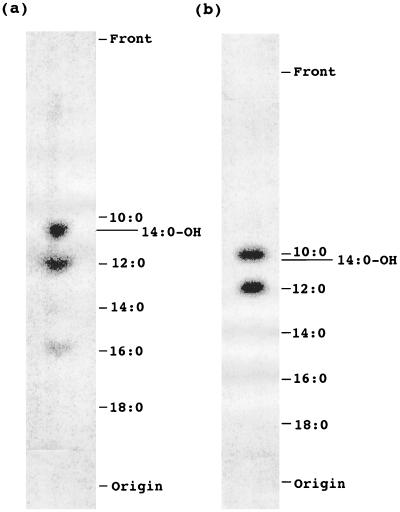
To further confirm that fatty acid synthesis takes place in mitochondria and to identify the products of the pathway, incubations were saponified, and the products labeled with [14C]malonate were methylated and then analyzed by reversed-phase and silica gel TLC. As shown in Fig. 2a, saponifiable radioactivity was mainly incorporated into hydroxymyristic (14:0-OH), lauric (12:0), and palmitic (16:0) acids. Because decanoic (10:0) and hydroxymyristic methyl esters are poorly resolved by reversed-phase TLC, the band comigrating with hydroxymyristic in Fig. 2a was recovered, and after TLC on silica gel H plates, 90% of the radioactivity comigrated with methyl hydroxymyristic and 10% with methyl decanoate (data not shown). The biosynthesis of hydroxymyristic is similar to the report that [14C]malonate is incorporated into 14:0-OH in mitochondria of Neurospora (15).
Figure 2.
Analysis of saponifiable fatty acids synthesized de novo in pea mitochondria with [2-14C]malonic acid. Fatty acids synthesized in pea mitochondria in vitro were separated on reversed-phase TLC plates and analyzed by PhosphorImager. (a) Total fatty acids. (b) Fatty acids bound to mtACP.
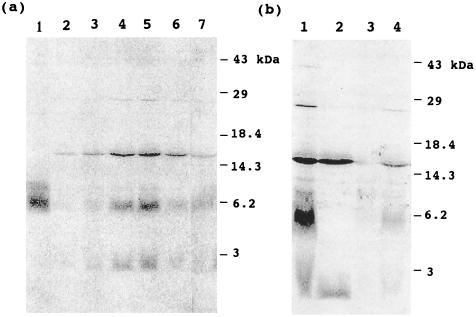
To determine if mtACP was involved in the fatty acid synthesis described above, 14C-labeled proteins were separated on SDS/PAGE and blotted onto nitrocellulose membrane, and radiolabeled proteins were detected by autoradiography (Fig. 3). Three major protein bands (which accounted for up to 50% of the 14C-labeled products of the incubation) were detected, and the radioactivity of each protein band increased with incubation time (Fig. 3a). The band at 6 kDa was identified as acyl-mtACP by comigration with the 14C-labeled 16:0-mtACP standard. Furthermore, the band at 6 kDa disappeared after DTT treatment under alkaline conditions, a treatment that cleaves the acyl groups bound to proteins by thioester bond (Fig. 3b, lane 2). These results indicate that the radioactive band at 6 kDa corresponds to acyl-mtACP. The other radioactive bands at 15 kDa and at <3 kDa were not altered by the DTT treatment, suggesting that acyl groups associated with these bands are not linked by thioester bonds.
Figure 3.
Labeling of pea mitochondrial proteins with [2-14C]malonic acid. Pea mitochondria were labeled with [2-14C]malonic acid, and proteins were separated by SDS/PAGE, blotted onto nitrocellulose membrane, and analyzed by PhosphorImager. (a) Time course of labeling of mitochondrial proteins and effect of ATP and MgCl2. In lanes 2–5, mitochondria were labeled for 10, 20, 40, and 60 min, respectively. In lanes 6 and 7, mitochondria were labeled for 1 h, but ATP or MgCl2, respectively, were omitted from assay medium. Lane 1 shows a standard of 14C-labeled 16:0-mtACP. (b) Effects of DTT treatment on deacylation of labeled proteins and cerulenin on labeling of proteins. Pea mitochondria were labeled for 1 h. Lane 1, control that was not treated with DTT; lane 2, mitochondrial proteins were treated with DTT; lanes 3 and 4, 200 μM and 20 μM cerulenin, respectively, were added to the incubation medium, and samples were not treated with DTT.
The 14C-labeled acyl groups bound to mtACP constituted ≈20% of the saponifiable fatty acids produced in these incubations and were analyzed by TLC after derivatization of acyl groups to FAMEs. As shown in Fig. 2b, mtACP was primarily acylated with 14C-labeled decanoic (10:0), lauric, and hydroxymyristic acids. Further analysis by silica gel TLC of the band migrating with decanoic indicated its composition as 80% decanoic and 20% hydroxymyristic (data not shown). Thus, the acyl-ACP pool includes more decanoic and less hydroxymyristic than the total saponifiable fatty acids (Fig. 2a).
To examine if the enzymes involved in fatty acid synthesis in mitochondria are similar to those of chloroplasts, the effect of cerulenin, which is an inhibitor for chloroplast fatty acid synthase-condensing enzymes, was investigated. As shown in Fig. 3b, cerulenin inhibited the labeling of proteins (lanes 3 and 4). This finding suggests that condensing enzymes involved in fatty acid synthesis in mitochondria are similar to those of chloroplasts. Furthermore, addition of ATP and Mg2+ to the incubations was essential for fatty acid synthesis and protein labeling (lanes 6 and 7).
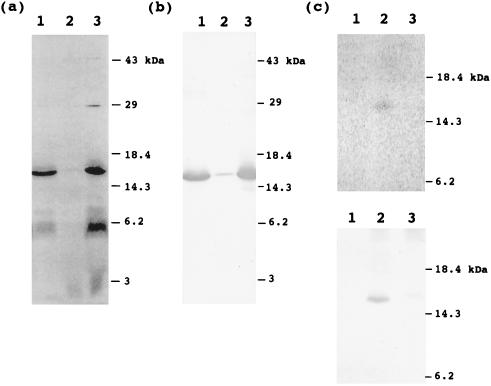
To investigate the localization of acyl-mtACP and other labeled proteins, the mitochondria labeled with [14C]malonate were fractionated into matrix and membrane fractions, and analyzed by SDS/PAGE. Fig. 4a shows the autoradiogram of proteins which were separated on SDS/PAGE and blotted to nitrocellulose membrane. It is clear that the acyl-mtACP and a 15-kDa protein are mainly localized in the matrix fraction, whereas the unidentified radioactivity migrating with molecular mass <3 kDa is mainly localized in the membrane fraction.
Figure 4.
Localization of 14C-labeled mitochondrial proteins. Pea mitochondria were fractionated into matrix and membrane fractions after 1 h of labeling with [2-14C]malonic acid. (a) Autoradiogram of proteins from each fraction. Proteins were separated by SDS/PAGE, blotted onto nitrocellulose membrane, and analyzed by PhosphorImager. Lane 1, matrix fraction; lane 2, membrane fraction; lane 3, mitochondria before fractionation. (b) Western blot analysis with an antibody against pea H protein. The same blot used for autoradiography in a was probed. Lanes 1–3, matrix fraction, membrane fraction, and mitochondria without fractionation, respectively. (c) Immunoprecipitation of H protein using an antibody against pea H protein. (Upper) Autoradiogram of immunoprecipitated proteins. Lanes 1–3, no IgG, 40 μg of immune IgG, and 60 μg of preimmune IgG were used for immunoprecipitation, respectively. (Lower) Western blot analysis of immunoprecipitated proteins. The same blot used for autoradiography was analyzed for Western blot analysis.
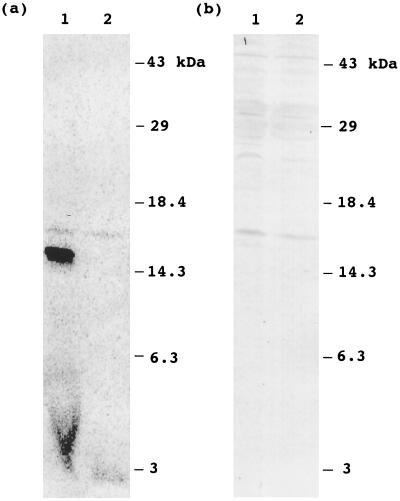
When the proteins in matrix and membrane fractions were separated on SDS/PAGE and were stained with Coomassie blue, we noticed that the location of the major radioactive band at 15 kDa comigrated with an abundant protein at 15 kDa. Over 30% of the total [14C]products produced in mitochondrial incubations could be attributed to this product. The 15-kDa size corresponds to that of H protein, which is a subunit of glycine decarboxylase and is an abundant protein located in the pea mitochondrial matrix. To verify that the labeled 15-kDa protein is H protein, the same blot used for the autoradiogram was probed with an antibody against pea H protein. As shown in Fig. 4b, a protein band identified by H protein antibodies was detected at a position identical to the radioactive band detected in the autoradiogram. Furthermore, the labeled 15-kDa protein could be immunoprecipitated with an antibody against pea H protein, but not with preimmune serum (Fig. 4c). Together, these findings demonstrate that the labeled 15-kDa protein is the H protein of glycine decarboxylase. To examine whether H protein is acylated in mitochondria of another eukaryote, mitochondria isolated from Neurospora were labeled with [14C]malonic acid, and the labeling of H protein was investigated. Fig. 5 shows an autoradiogram of labeled proteins from Neurospora and the result of Western blot analysis with an antibody against pea H protein. Two radioactive protein bands appeared in the autoradiogram. The major band at 15 kDa has an apparent molecular mass similar to that reported for Neurospora acyl-mtACP (2, 15, 27), and this band disappeared after treatment with DTT under alkaline conditions. These findings suggest that this band corresponds to acyl-mtACP. The minor band at 16 kDa did not disappear upon the DTT treatment, indicating radioactivity is not linked to the protein via a thioester. When the same blot was probed with antibodies to pea H protein, a crossreactive band was detected at the identical position to the 16-kDa radioactive band. These findings demonstrate that H protein is acylated not only in pea mitochondria, but also in Neurospora mitochondria. The higher radioactivity in the pea H protein, compared with that of Neurospora, is consistent with the much higher content of glycine decarboxylase in pea leaf mitochondria, where it accounts for up to 50% of the matrix proteins.
Figure 5.
Labeling of Neurospora mitochondria with [2-14C]malonic acid. Neurospora mitochondria were labeled with [2-14C]malonic acid for 1 h. Proteins without DTT treatment or proteins treated with DTT were separated by SDS/PAGE, blotted onto nitrocellulose membrane, and analyzed by PhosphorImager. (a) Autoradiogram of mitochondrial proteins. Lanes 1 and 2, proteins without DTT treatment and the proteins treated with DTT, respectively. (b) Western blot analysis of mitochondrial proteins probed with antibodies to pea H protein. Lanes 1 and 2, proteins without DTT treatment and the proteins treated with DTT, respectively. The same blot used for autoradiography in a was used for Western blot analysis.
It is known that H protein contains lipoic acid as an essential cofactor and that lipoic acid is bound to a lysine residue by an amide bond (28–30). This raised the possibility that the H protein is acylated with lipoic acid synthesized from fatty acids in pea mitochondria. Because an amide linkage is stable to saponification, the analysis of fatty acid products shown in Fig. 2 would not have included amide-linked acyl groups. The identity of the radioactive acyl group attached to pea H protein was determined by cleavage of the amide linkage with 6 M HCl. Over 70% of the radioactivity released from H protein comigrated with octanoic acid on reversed-phase TLC (data not shown). This result is similar to data obtained with E. coli, where octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex takes place in a lipoic acid-auxotrophic mutant (31, 32). Thus, in these isolated mitochondria, the complete pathway for lipoic acid production could not be demonstrated and may depend on additional factors not present in our incubations. However, the mitochondrial extracts of pea and Neurospora prepared for this study catalyzed transfer of lipoate from lipoyl-ACP to the E2 subunit of E. coli apo-pyruvate dehydrogenase (J. E. Cronan, Jr., and S. Jordan, personal communication) further confirming the presence of a lipoyl-cofactor assembly pathway in mitochondria.
DISCUSSION
The function of fatty acid biosynthesis in mitochondria has remained an enigma. In eukaryotic plants, synthesis of fatty acids occurs primarily in plastids such as chloroplasts, and products of this pathway supply acyl precursors for mitochondrial and other extraplastidial membranes. The discovery of ACP in plant mitochondria (4, 5) has raised the possibility that these organelles also participate in de novo fatty acid synthesis. However, ACP has been considered to have several other functions in metabolism, and, in mitochondria, ACP is reported to be one component of the respiratory electron transport chain (14). Thus, whether ACP participates in fatty acid synthesis in plant mitochondria and what function this pathway might have in any eukaryotic organism has been unclear. In this study, we have demonstrated that the de novo synthesis of fatty acids does occur in pea mitochondria, indicating that these organelles possess the complete set of enzymes needed to assemble fatty acids. The analysis of fatty acids synthesized in mitochondria demonstrated that the major products of the pathway in pea mitochondria were (i) octanoic acid attached to the H protein of glycine decarboxylase and (ii) hydroxymyristic, lauric, and palmitic fatty acids. These products are strikingly different from the predominantly 18-carbon fatty acids synthesized in chloroplasts or present in plant membranes and demonstrate that an alternative biosynthetic pathway for fatty acids is present in mitochondria of higher plants.
It is known that H protein, a subunit of glycine decarboxylase, contains lipoic acid as an essential cofactor (33). The biosynthesis of lipoic acid has been best studied in E. coli by molecular genetic approaches (32, 34–37). Three genes, which are involved in lipoic acid metabolism, have been cloned from E. coli by complementation of mutants defective in lipoic acid biosynthesis. It has been clarified that lipoic acid is synthesized from octanoyl-ACP via the insertion of two sulfur atoms into an octanoyl group to produce a dithiolane ring. One of the genes, lplA, encodes a lipoyl-protein ligase for the attachment of lipoyl groups to lipoate-dependent apoenzymes (36). The other genes, lipA and lipB, encode an enzyme involved in the formation of the dithiolane ring from an octanoyl group and probably another lipoyl-protein ligase, respectively (32, 34). By contrast, the biosynthesis of lipoic acid in eukaryotes has not been well studied. It is not known how lipoic acid is synthesized nor where the enzymes involved in the biosynthesis of lipoic acid are localized in eukaryotes. Fujiwara et al. (38) observed that H protein is lipoylated in mitochondria when the precursor of H protein was imported into mitochondria in vitro. This finding suggests that the H protein is lipoylated after the transport of the precursor into mitochondria and therefore lipoic acid may be synthesized in mitochondria. Sulo and Martin (39) isolated a LIP5 gene from S. cerevisiae by complementation of a mutant defective in lipoic acid synthesis. The protein sequence deduced from the gene is homologous to the product of the E. coli lipA gene. Relative to the E. coli gene product, the LIP5 protein has an amino-terminal extension with characteristics of mitochondrial targeting signals. These findings also indicate that the biosynthesis of lipoic acid in eukaryotes occurs in mitochondria.
In the present study, when pea mitochondria were labeled with [2-14C]malonate, we observed that H protein was highly labeled, accompanied by the acylation of mtACP. The acylations of mtACP and H protein were also observed in mitochondria isolated from Neurospora. These findings indicate that lipoic acid is synthesized from a fatty acid precursor produced in mitochondria and that a major function of mtACP is the biosynthesis of lipoic acid. Although the incorporation of sulfur into octanoic acid to form lipoic acid is still not well understood, the findings obtained in this study provide a new understanding of the biosynthetic pathway for the octanoyl precursor of lipoic acid in eukaryotes and of the function of mitochondrial fatty acid synthesis.
Acknowledgments
We thank Dr. Helmut Bertrand (Michigan State University) for the generous gift of mitochondria of Neurospora. We also gratefully acknowledge Dr. David J. Oliver (Iowa State University) for providing the antibody against pea H protein. This work was supported by a grant from the National Science Foundation (DCB90-05290). We also acknowledge the Michigan Agricultural Experiment Station for its support of this research.
Footnotes
Abbreviations: ACP, acyl carrier protein; mtACP, mitochondrial ACP; FAME, fatty acid methyl ester; X, number of carbon atoms in a fatty acid; Y, number of double bonds in a fatty acid.
References
- 1.Brody S, Mikolajczyk S. Eur J Biochem. 1988;173:353–359. doi: 10.1111/j.1432-1033.1988.tb14005.x. [DOI] [PubMed] [Google Scholar]
- 2.Sackmann U, Zensen R, Rohlen D, Jahnke U, Weiss H. Eur J Biochem. 1991;200:463–469. doi: 10.1111/j.1432-1033.1991.tb16205.x. [DOI] [PubMed] [Google Scholar]
- 3.Schneider R, Massow M, Lisowsky T, Weiss H. Curr Genet. 1995;29:10–17. doi: 10.1007/BF00313188. [DOI] [PubMed] [Google Scholar]
- 4.Chuman L, Brody S. Eur J Biochem. 1989;184:643–649. doi: 10.1111/j.1432-1033.1989.tb15061.x. [DOI] [PubMed] [Google Scholar]
- 5.Shintani D K, Ohlrogge J B. Plant Physiol. 1994;104:1221–1229. doi: 10.1104/pp.104.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Runswick M J, Fearnley I M, Skehel J M, Walker J E. FEBS Lett. 1991;286:121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- 7.Ohlrogge J B. In: The Biochemistry of Plants. Stumpf P K, Conn E E, editors. Vol. 9. New York: Academic; 1987. pp. 137–157. [Google Scholar]
- 8.Ohlrogge J, Browse J. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlrogge J B, Kuhn D N, Stumpf P K. Proc Natl Acad Sci USA. 1979;76:1194–1198. doi: 10.1073/pnas.76.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakil S J, Stoops J K, Joshi V C. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 11.Therisod H, Weissborn A C, Kennedy E P. Proc Natl Acad Sci USA. 1986;83:7236–7240. doi: 10.1073/pnas.83.19.7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen B, Summers R G, Gramajo H, Bibb M J, Hutchinson C R. J Bacteriol. 1992;174:3818–3821. doi: 10.1128/jb.174.11.3818-3821.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issartel J-P, Koronakis V, Hughes C. Nature (London) 1991;351:759–761. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 14.Schulte U, Fecke W, Krüll C, Nehl U, Schmiede A, Schneider R, Ohnishi T, Weiss H. Biochim Biophys Acta. 1994;1187:121–124. doi: 10.1016/0005-2728(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 15.Mikolajczyk S, Brody S. Eur J Biochem. 1990;187:431–437. doi: 10.1111/j.1432-1033.1990.tb15322.x. [DOI] [PubMed] [Google Scholar]
- 16.Arnon D I. Plant Physiol. 1949;64:182–186. [Google Scholar]
- 17.Lizardi P M, Luck D J. Nature (London) New Biol. 1971;229:140–142. doi: 10.1038/newbio229140a0. [DOI] [PubMed] [Google Scholar]
- 18.Lambowitz A M. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- 19.Infanger A, Bertrand H. Curr Genet. 1986;10:607–617. doi: 10.1007/BF00418128. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Battey J F, Ohlrogge J B. Planta. 1990;180:352–360. doi: 10.1007/BF00198786. [DOI] [PubMed] [Google Scholar]
- 22.Kuo T M, Ohlrogge J B. Arch Biochem Biophys. 1984;230:110–116. doi: 10.1016/0003-9861(84)90091-2. [DOI] [PubMed] [Google Scholar]
- 23.Post-Beittenmiller, Jaworski J G, Ohlrogge J B. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- 24.Roesler K R, Savage L J, Shintani D K, Shorrosh B S, Ohlrogge J B. Planta. 1996;198:517–525. doi: 10.1007/BF00262637. [DOI] [PubMed] [Google Scholar]
- 25.Baldet P, Alban C, Axiotis S, Douce R. Arch Biochem Biophys. 1993;303:67–73. doi: 10.1006/abbi.1993.1256. [DOI] [PubMed] [Google Scholar]
- 26.Roughan P G. Methods Enzymol. 1987;148:327–337. [Google Scholar]
- 27.Zensen R, Husmann H, Schneider R, Peine T, Weiss H. FEBS Lett. 1992;310:179–181. doi: 10.1016/0014-5793(92)81324-f. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara K, Okamura-Ikeda K, Motokawa Y. J Biol Chem. 1986;261:8836–8841. [PubMed] [Google Scholar]
- 29.Kim Y, Oliver D J. J Biol Chem. 1990;265:848–853. [PubMed] [Google Scholar]
- 30.Macherel D, Lebrun M, Gagnon J, Neuburger M, Douce R. Biochem J. 1990;268:783–789. doi: 10.1042/bj2680783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali S T, Moir A J G, Ashton P R, Engel P C, Boom T J V, Reed K E, Cronan J E., Jr J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris T W, Reed K E, Cronan J E., Jr J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara K, Okamura K, Motokawa Y. Arch Biochem Biophys. 1979;197:454–462. doi: 10.1016/0003-9861(79)90267-4. [DOI] [PubMed] [Google Scholar]
- 34.Boom T J V, Reed K, Cronan J E., Jr J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed K E, Cronan J E., Jr J Bacteriol. 1993;175:1325–1336. doi: 10.1128/jb.175.5.1325-1336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris T W, Reed K E, Cronan J E., Jr J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- 37.Reed K E, Morris T W, Cronan J E., Jr Proc Natl Acad Sci USA. 1994;91:3270–3724. doi: 10.1073/pnas.91.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujiwara K, Okamura-Ikeda K, Motokawa Y. J Biol Chem. 1990;265:17463–17467. [PubMed] [Google Scholar]
- 39.Sulo P, Martin N C. J Biol Chem. 1993;268:17634–17639. [PubMed] [Google Scholar]