Abstract
Depudecin is a fungal metabolite that reverts the rounded phenotype of NIH 3T3 fibroblasts transformed with v-ras and v-src oncogenes to the flattened phenotype of the nontransformed parental cells. The mechanism of detransformation induced by this agent had not been determined. Here, we demonstrate that depudecin inhibits histone deacetylase (HDAC) activity effectively both in vivo and in vitro. Depudecin induces similar morphological reversion in v-ras transformed NIH 3T3 cells as do other naturally occurring HDAC inhibitors such as trichostatin A or trapoxin. It competitively inhibits the binding of [3H]trapoxin in vitro and the nuclear binding of a trapoxin–coumarin fluorophore in vivo, suggesting that depudecin shares a nuclear binding protein and site on that protein with trapoxin. Furthermore, depudecin induces hyperacetylation of histones in a dose-dependent manner and at concentrations comparable with that required for detransformation. An in vitro histone deacetylase assay, using purified recombinant HDAC1, reveals that depudecin inhibits 50% of the enzyme activity at a concentration of 4.7 μM. These results demonstrate that depudecin is a novel HDAC inhibitor and suggest that its ability to induce morphological reversion of transformed cells is the result of its HDAC inhibitory activity.
The shape and cytoskeletal architecture of cells are often dramatically altered as a consequence of malignant transformation (1). One of the most characteristic cytoskeletal changes in tumor cells is the loss of actin stress fibers, suggesting that the actin-containing microfilament system is a critical target during tumorigenesis (2). Agents that reorganize these fibers, reverting the transformed morphology to a normal one, can be used to probe growth factor-mediated signaling pathways leading to actin stress fiber formation, and have potential as therapeutics for treatment of cancer.
Several natural products have been isolated by screening for detransforming activity using oncogene-transformed cells. Examples include trapoxin (3, 4), trichostatin A (5), herbimycin A (6), radicicol (7), and azatyrosine (8). These compounds are able to revert the morphological changes seen following the transformation of cells in culture with v-sis, v-src, or v-ras oncogenes and in cells derived from tumors. For instance, trichostatin A and trapoxin are known to revert v-sis and v-ras oncogene-transformed cells as well as human tumor cells such as HeLa and T24 cells (9, 10). It has been unclear how these compounds are able to show a broad spectrum of detransforming activity in tumor cells. Histone deacetylases (HDACs) were considered likely molecular targets of these agents (11), and, recently, human histone deacetylase (HDAC1) was purified and cloned by using a trapoxin-based affinity matrix purification (12). Accumulating evidence suggests that the acetylation and deacetylation of histones play significant roles in the regulation of transcription in eukaryotic cells (13–16). The broad spectrum of detransforming activities displayed by HDAC inhibitors implies a role for HDACs in stress fiber formation and cell growth control (9, 11). HDACs may repress the transcription of anti-tumor genes whose products induce actin stress fiber reorganization and arrest cell proliferation.
Trichostatin A and trapoxin are the only detransforming agents known to target HDACs. For example, radicicol, shown here not to inhibit HDAC enzymatic activity or trapoxin binding to HDACs, is thought to mediate its actions by inhibiting Src tyrosine kinase activity (7). Depudecin (17) is a structurally novel natural product whose molecular target had not previously been defined (18). It was discovered in the culture broth of the fungus Alternaria brassicicola by assaying for the morphological detransformation of NIH 3T3 cells doubly transfected with v-ras and v-src oncogenes. Depudecin does not suppress the expression of the ras gene, suggesting that its mode of action does not relate directly to Ras function (19). It also shows a spectrum of detransforming activity, both in v-raf transformed cells and in a human osteosarcoma cell line, MG63 (18).
In this study, we demonstrate that depudecin inhibits histone deacetylase activity both in vivo and in vitro. Depudecin represents the fourth structural class of molecules (see Fig. 1) having this activity. Like the other three, its inhibitory activity can be rationalized in terms of its unique chemical structure and the anticipated structural features of the substrate binding and catalytic sites on HDAC enzymes. Like the actin and tubulin proteins, which are liganded by many different microbial products having diverse chemical structures, HDACs seem to have been targeted frequently by microbial biosynthetic pathways.
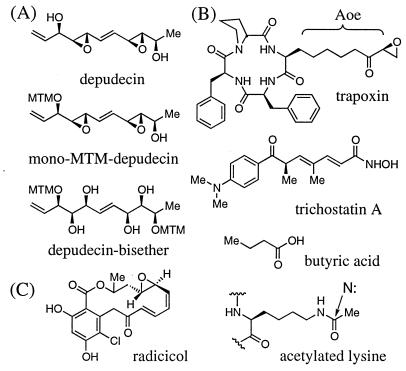
Figure 1.
The chemical structures of depudecin and other compounds discussed in text. (A) Depudecin and its inactive derivatives, mono-MTM-depudecin and depudecin-bisether. (B) HDAC inhibitors trapoxin, trichostatin, and butyric acid. (C) Radicicol. Trapoxin and other cyclic tetrapeptide inhibitors contain the functionally important amino acid (2S,9S)-2-amino-8-oxo-9,10-epoxydecanoic acid (Aoe). This side chain is approximately isosteric to that of ɛ-N-acetylated lysine residues in histone proteins and may form a bond to a nucleophilic residue in the enzyme active site.
MATERIALS AND METHODS
Materials.
Depudecin and its inactive derivatives used in this study were prepared by total synthesis according to the method of Shimada et al. (18). [3H]Trapoxin was prepared by total synthesis as described by Taunton et al. (20). Trichostatin A was purchased from Biomol (Plymouth Meeting, PA), and radicicol was provided generously as a gift by Professor W. C. Taylor (University of Sydney). DMEM, RPMI media, fetal calf serum, glutamine, and penicillin-streptomycin were purchased from GIBCO/BRL.
Synthesis of a Trapoxin–Coumarin Fluorophore.
An allyloxycarbonyl-protected, lysine variant of trapoxin, previously named “K-trap,” was prepared as described by Taunton et al. (20). The allyloxycarbonyl group was removed from K-trap by brief treatment with 10 mol% of tris(dibenzylideneacetone) dipalladium chloroform and 40 mol% of triphenylphosphine in the presence of dimedone (5 equivalent). The crude amine was purified immediately by reverse phase HPLC, lyophilized, and coupled to N-hydroxysuccinimide ester of a coumarin dye (Molecular Probes) in the presence of excess N,N-diisopropylethylamine. The reaction mixture was treated with saturated NaHCO3, extracted with EtOAc, dried by filtration through anhydrous MgSO4, and concentrated in vacuo. The crude product was purified by small pipet column chromatography to afford a trapoxin–coumarin conjugate, named “K-trap–coumarin dye” in Fig. 4 (molecular weight 898.07) in 65% combined yield for two steps. Its molecular weight and chemical structure were confirmed using fast atom bombardment-MS and 1H-NMR.
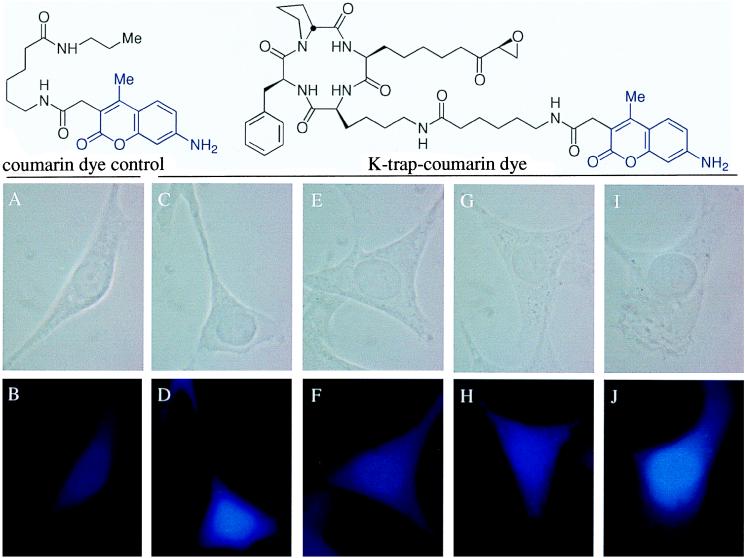
Figure 4.
Visualization of HDACs and HDAC binding in the nucleus of v-ras NIH 3T3 cells using a competition binding assay involving K-trap–coumarin dye and various detransformation agents. The staining was carried out as described in Material and Methods. Upper and lower photos were taken using phase-contrast and fluorescence microscopy, respectively. Coumarin dye excites at 345 nm with blue light. (A and B) Coumarin dye control. (C and D) K-trap–coumarin dye control. (E and F) K-trap–coumarin dye in cells pretreated with 23.5 μM depudecin. (G and H) K-trap–coumarin dye in cells pretreated with 0.1 μM trapoxin. (I and J) K-trap–coumarin dye in cells pretreated with 3.4 μM radicicol.
Cells and Cell Culture Conditions.
v-ras transformed NIH 3T3 cells were kindly supplied by Professor R. L. Erickson (Molecular and Cellular Biology, Harvard University), and HL60 and COS cells were purchased from ATCC. These cells were cultured at 37°C in a humidified atmosphere of 5% CO2 in DMEM (v-ras NIH 3T3 and COS) or RPMI (HL60) medium supplemented with 10% fetal calf serum, glutamine, and penicillin-streptomycin.
Cell Staining with a Trapoxin–Coumarin Fluorophore.
To obtain evidence of in vivo binding of depudecin to nuclear HDACs, cells were stained with the trapoxin–coumarin fluorophore in analogy to the method of Shimada et al. (18). Briefly, asynchronous cells were cultured on a slide coverslip for 18 h. After treatment with depudecin or other inhibitors for an additional 24 h, cells were fixed with 3.7% paraformaldehyde in PBS for 20 min at room temperature. Following three times washing with PBS, cells were treated with 0.1% Triton X-100 in PBS for 5 min to increase cell permeability. Next, the cells were treated with the trapoxin–coumarin fluorophore (5 μg) for 20 min, and after washing three times with PBS, the cells were mounted on a slide with an antifader (DABCO, 1, 4-diazabicyclo[2.2.2]-octane, Aldrich) in 50% glycerol. Cells were inspected by conventional fluorescence microscopy (Leitz Wetzlar, Germany).
[3H]Trapoxin Binding Assay.
Soluble extracts for binding studies were prepared by homogenizing 1 × 108 v-ras NIH 3T3 or HL60 cells in 50 mM Tris-HCl (pH 7.5) with 100 mM NaCl and 10% glycerol. The resulting homogenate was centrifuged in a microcentrifuge at maximum speed (15,000 rpm) for 15 min, and the supernatant was used directly. For the competition binding assay with [3H]trapoxin, test compounds were preincubated with the supernatant (200 μg/100 μl) for 1 h at 25°C, and then 100 nM [3H]trapoxin was added for 30 min at 25°C. Free radioactivity was removed by precipitation with 1% charcoal solution in 10% BSA, and the binding activity was determined by scintillation counting of the supernatant.
Histone Hyperacetylation Assay.
Histones were extracted from v-ras NIH 3T3 or HL60 cells (2 × 107 cells) that had been treated with test compounds for 6 h at 37°C as described by Yoshida et al. (21). Acid urea gel electrophoresis was used for the detection of acetylated histone molecules. Slab gels were run at 12 mA for 10 h at room temperature as described by Panyim and Chalkley (22) using a slab gel.
Histone Deacetylase Assays.
In vitro histone deacetylase activity was assayed as described previously with either 100 μl of crude cell extract (1 × 107 cells) or 25 ng of recombinant FLAG epitope-tagged HDAC1 (HDAC1-F) for 3 h at 37°C (refs. 12 and 23; C.A.H., J. K. Tong, T.O., D. E. Ayer, and S.L.S., unpublished results). Pretreatment of crude or HDAC1-F enzymes with test compounds was performed for 30 min at 4°C prior to addition of the [3H]acetyl histone substrate that was acetylated with [3H]acetate (0.5 Ci/mmol, New England Nuclear) by an in vivo incorporation method (21). The reaction was quenched with 1 M HCl and 0.16 M acetic acid (50 μl). Released [3H]acetate was extracted with 600 μl of ethyl acetate and quantified by scintillation counting.
RESULTS
Induction of Morphological Change in v-ras NIH 3T3 Cells by Depudecin.
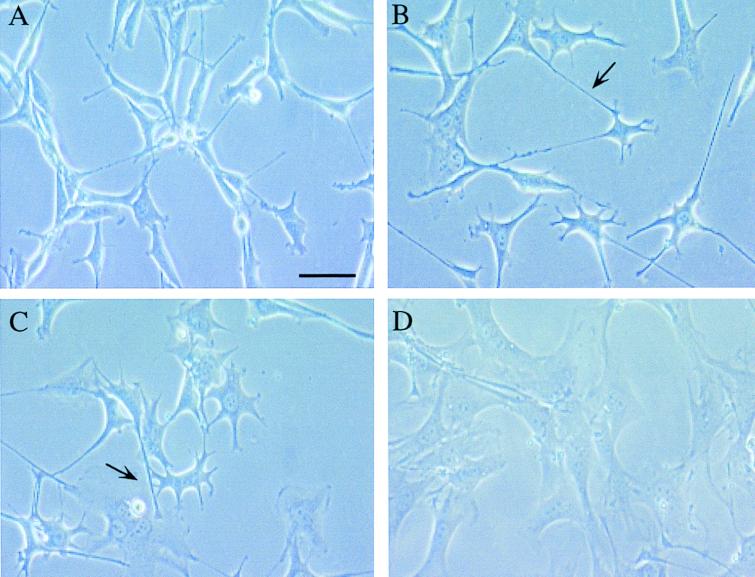
v-ras NIH 3T3 is a cell line that is known to revert from a spindle-like, transformed to a normal morphology following treatment with depudecin, trichostatin A, or radicicol. We first compared the effects of these agents on cell morphology. After treatment for 24 h, the cells showed pronounced and characteristic morphological alterations depending on the agent (Fig. 2). Generally, the spindle-like morphology of v-ras NIH 3T3 cells was changed to a flattened morphology by all of these compounds, as reported previously (9, 18, 24). However, cells treated with depudecin and trichostatin A produced characteristic elongated cells with filamentous protrusions (noted by arrows in Fig. 2), whereas the tyrosine kinase inhibitor radicicol did not. Trapoxin, another HDAC inhibitor, induced morphologic alterations reminiscent of those induced by depudecin and trichostatin A (data not shown), suggesting that the detransforming activity of depudecin may also be due to the inhibition of HDAC enzymes.
Figure 2.
The morphological change in v-ras-transformed NIH 3T3 cells by detransforming agents. Exponentially growing cells were treated for 24 h with test agents, and photographs were taken under a phase-contrast microscope. (A) Control v-ras transformed NIH 3T3 cells. (B) Cells treated with 4.7 μM synthetic depudecin. (C) Cells treated with 1.5 μM trichostatin A. (D) cells treated with 3.4 μM radicicol. The characteristic elongated cells having filamentous protrusions induced by depudecin and trichostatin A are indicated by the arrow. (Bar = 50 μm.)
Depudecin Binds Competitively to a [3H]Trapoxin-Binding Protein.
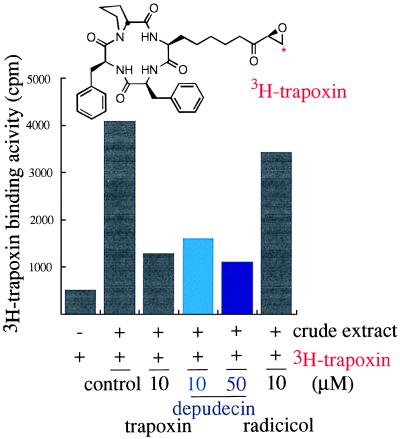
To explore HDAC inhibition as the potential mode of action of depudecin in detransformation, a binding assay was carried out to see whether depudecin shares a binding protein with known HDAC inhibitors. [3H]Trapoxin was chosen as the radioligand because it shows a high specific binding to HDACs (20) (Fig. 3). Depudecin, like trapoxin, also effectively inhibited [3H]trapoxin binding in a dose-dependent manner. However, radicicol was largely ineffective as a competitive inhibitor of [3H]trapoxin binding. These results indicate that depudecin shares common target proteins with trapoxin, likely the HDAC enzymes, and that radicicol does not.
Figure 3.
The effect of depudecin and other detransforming agents on [3H]trapoxin binding with crude lysate of v-ras transformed NIH 3T3 cells. The assay was carried out as described in Material and Methods. A 100-fold excess amount of cold compounds was preincubated with crude lysate for 30 min at 25°C in the competition assay.
Depudecin Prevents Appearance of Fluorescence in the Nucleus Induced by a Trapoxin–Coumarin Dye.
To determine whether depudecin binds to HDACs in v-ras NIH 3T3 and COS cells, we synthesized a fluorescent variant of trapoxin, named K-trap–coumarin dye (Fig. 4). This reagent inhibited HDAC activity at a concentration similar to that of K-trap (100 nM) both in vitro and in vivo, suggesting that K-trap–coumarin dye binds HDACs and should therefore be competed by the pretreatment of other HDAC inhibitors. As shown in Fig. 4, K-trap–coumarin dye stains the nuclei (blue color), whereas coumarin dye control does not show a significant signal (Fig. 4 A and B, C and D, respectively). This result provides evidence for the expected nuclear localization of HDAC proteins. When cells were pretreated with depudecin or trapoxin, similar morphological changes (i.e., spread and elongated protrusion morphology) were observed, but the fluorescence signal was weaker and distributed throughout the cell (Fig. 4 E and F, G and H, respectively). This result suggests that depudecin competes with K-trap–coumarin dye for binding to HDACs in cells, as does trapoxin. Interestingly, cells treated with radicicol show an altered (flattened) morphology, but the K-trap–coumarin dye stains the nuclei to comparable levels as the control, suggesting that radicicol induces a morphological change by a mechanism not involving HDAC binding (Fig. 4 I and J). Depudecin-bisether, an inactive depudecin derivative, does not change the morphology of cells nor inhibit the K-trap–coumarin dye from staining the nuclei (data not shown).
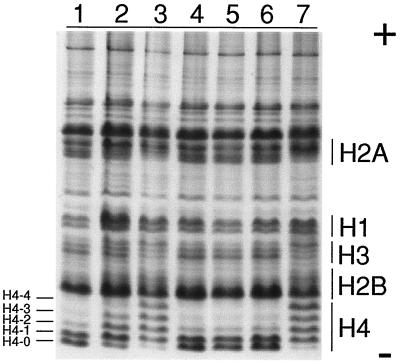
Depudecin Induces Histone Hyperacetylation in Vivo.
We next investigated whether depudecin can inhibit histone deacetylase activity in v-ras NIH 3T3 cells. It is known that inhibition of this activity by trichostatin A (21), trapoxin (10), and sodium butyrate (25) results in the accumulation of hyperacetylated histone species. This effect can be visualized by acid/urea/TritonX-100 (AUT) gel electrophoresis of histones extracted from inhibitor-treated cells. Increased acetylation results in a decreased mobility of histones in AUT gel electrophoresis and the appearance of multiple bands corresponding to histones with 1, 2, 3, and 4 acetylated residues. As shown in Fig. 5, depudecin induces hyperacetylation of histones (especially histone H4) in a dose-dependent manner. Trichostatin A, but not radicicol, also strongly induces hyperacetylation of histones. We had previously reported that mono-methylthiomethyl (MTM)-depudecin and depudecin-bisether, two inactive depudecin derivatives, did not induce detransformation in v-ras NIH 3T3 cells (18). These inactive depudecin derivatives do not induce accumulation of acetylated histones (Fig. 5, lanes 4 and 5). These results demonstrate that depudecin inhibits HDAC activity in vivo and suggest that depudecin’s detransforming activity may be manifested via HDAC inhibition.
Figure 5.
The effect of depudecin and other detransforming agents on histone acetylation in v-ras transformed NIH 3T3 cells. Lane 1, control, treatment of v-ras transformed NIH 3T3 cells for 6 h with vehicle only; lane 2, 4.7 μM depudecin; lane 3, 23.5 μM depudecin; lane 4, 47 μM mono-MTM-depudecin; lane 5, 47 μM depudecin-bisether; lane 6, 3.4 μM radicicol; lane 7, 1.5 μM trichostatin A. Histones were extracted from the cells and visualized on AUT gel by Coomassie brilliant blue staining. Acetylated histone species have slower rates of migration relative to nonacetylated histones (H4-0, -1, -2, -3, -4).
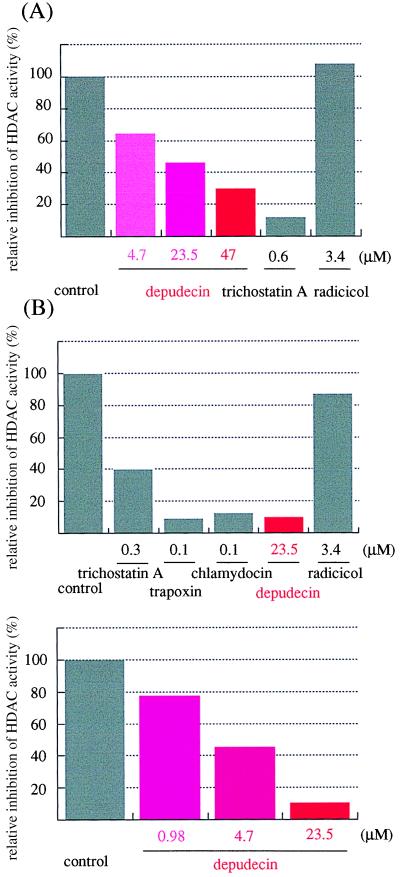
Depudecin Inhibits HDAC Activity in Vitro.
Next, we confirmed the inhibitory activity of depudecin on purified HDACs in vitro. First, we used an extract from HL60 cells as a source of HDACs. Extracts were incubated with several detransforming agents and examined for HDAC activity using 3H-acetylated histones as substrate. Depudecin inhibited the HDAC activity in a dose-dependent manner. Trichostatin A also showed strong inhibition of the enzyme, whereas radicicol did not (Fig. 6A). We next investigated the effect of depudecin on purified FLAG epitope-tagged HDAC1 (HDAC1-F) expressed in Sf9 insect cells using a baculovirus vector (23). As shown at Fig. 6B, depudecin inhibited the purified enzyme activity to levels comparable with the known HDAC inhibitors, trichostatin A, trapoxin, and chlamydocin (26). In contrast, radicicol had a negligible effect. Higher concentration (47 μM) of the inactive depudecin-bisether did reduce activity of pure HDAC1-F but not HDAC activity of an extract (data not shown). The IC50 value of depudecin in this assay (4.7 μM) is comparable with that determined in the detransforming assay (4.7–47 μM) (19).
Figure 6.
The effect of depudecin and other detransforming agents on histone deacetylase in vitro. The assay was carried out as described in Materials and Methods. (A) HDAC assay using crude lysate as the source of enzyme. (B) HDAC assay using purified recombinant HDAC1-F.
DISCUSSION
Depudecin is a natural product having a highly unusual structure. Its 11-carbon chain contains two epoxides and six stereogenic centers. Until now, the mechanism of its ability to induce actin stress fiber formation in cells was unknown. In part by synthesizing a fluorescent HDAC ligand and using it as a visual reporter of nuclear HDAC binding, the present study demonstrates that depudecin inhibits HDAC activity both in vitro and in vivo. Because other, structurally unrelated HDAC inhibitors have similar effects on cells, including the ability to induce actin stress fibers, the results suggest that HDAC enzymes are the biologically relevant target proteins of depudecin. However, the present study also shows that at least one other inducer of actin stress fiber formation, radicicol, does not operate through an HDAC mechanism. Depudecin and its derivatives should be useful tools to explore the role of HDACs and histone acetyltransferases (27) in the regulation of chromatin structure and gene transcription in eukaryotes (23, 28–31).
The molecular basis for the downstream effects of HDAC inhibition by depudecin remains to be determined. Depudecin induces not only morphological changes but also cell cycle arrest and cellular differentiation in many mammalian cell lines (H.J.K. and S.L.S., unpublished results), as do other HDAC inhibitors. The detransforming activity of depudecin is suppressed by actinomycin D and cycloheximide, suggesting that both mRNA synthesis and de novo protein synthesis are required for depudecin-mediated detransforming activity (18). HDAC inhibitors result in increased levels of gelsolin, a Ca2+-dependent actin filament-severing and capping protein, likely by derepression of the gelsolin gene (9, 32). That this effect is relevant to the observed morphological changes is supported by the observation that HDAC inhibitor-induced morphologic changes are suppressed following microinjection of anti-gelsolin antibodies (33).
Several HDAC inhibitors have been used to explore the function of HDACs in cells. The earliest identified HDAC inhibitor, butyric acid, inhibits HDAC enzymatic activity at millimolar concentration, leading to concerns that this agent induces additional effects unrelated to histone hyperacetylation. The potency and apparent specificity of the natural product inhibitors, products of natural selection, largely alleviate these concerns. That nature has produced a wide variety of structures from a variety of sources for the purpose of HDAC inhibition suggests that these enzymes play essential and conserved functions. Two other essential proteins that have been targeted by this form of natural selection are actin and tubulin.
The mechanism of HDAC inhibition by depudecin may be related to that of the other natural inhibitors, especially that of trapoxin. Each of these compounds has an acyclic chain presumed to mimic the side chain of an ɛ-N-acetylated lysine residue. At the termini of these chains are different chemical moieties that likely interact with key catalytic elements in the enzyme active sites. The carboxylic and hydroxamic acid groups of butyrate and trichostatin most likely ligand a catalytic metal ion. The epoxide groups in the depudecin, trapoxin, and trapoxin-related natural products (e.g., chlamydocin) likely bond covalently with nucleophilic groups in the active sites. The selectivity of these inhibitors toward different HDAC family members is not currently known (12, 34, 35).
Depudecin and other HDAC inhibitors have potential as therapeutic agents. Apicidin, a cyclic peptide related to trapoxin, has been shown to exhibit potent antiprotozoal activity by inhibition of HDACs in parasites (36). Depudecin has been shown to exhibit anti-angiogenesis activity both in vitro and in vivo (37), suggesting that specific inhibition of HDACs in endothelial cells could have therapeutic value. HDAC inhibitors arrest the cell cycle and revert the transformed morphology of p53-deficient cell lines HL60, HeLa, Saos-2, and of SV40-transformed cells (11), suggesting a strategy for treatment of tumors deficient in p53 function (38). Future investigations of HDAC function and HDAC-related therapies will benefit from variants of the natural inhibitors showing specificity for individual HDAC family members and for HDACs from specific organisms. Their structures, now including that of depudecin, combined with new methods in small molecule synthesis and screening, will provide a useful guide to such research efforts.
Acknowledgments
We thank H. Nakajima and J. K. Tong for useful discussions and M. Foley for his assistance in the synthesis of depudecin. T.O. and J.S. are visiting scientists from Eisai Co., Ltd. and Kyowa Hakko Kogyo Co., Ltd., respectively. C.A.H. is supported by a National Institutes of Health predoctoral training grant, and H.J.K. and S.L.S. are a Research Associate and Investigator, respectively, at the Howard Hughes Medical Institute.
ABBREVIATIONS
- HDAC
histone deacetylase
- AUT
acid/urea/Triton X-100
Footnotes
A commentary on this article begins on page 3335.
References
- 1.Wahrman M Z, Gagnier S E, Kobrin D R, Higgins P J, Augenlicht L H. Tumor Biol. 1985;6:41–56. [PubMed] [Google Scholar]
- 2.Wang Y-I, Goldberg A R. Proc Natl Acad Sci USA. 1976;73:4065–4069. doi: 10.1073/pnas.73.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itazaki H, Nagashima K, Sugita K, Yoshida H, Kawamura Y, Yasuda Y, Matsumoto K, Ishii K, Uotani N, Nakai H, Terui A, Yoshimatsu S, Ikenishi Y, Nakagawa Y. J Antibiot. 1990;43:1524–1532. doi: 10.7164/antibiotics.43.1524. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H, Sugita K. Jpn J Cancer Res. 1992;83:324–328. doi: 10.1111/j.1349-7006.1992.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugita K, Koizumi K, Yoshida H. Cancer Res. 1992;52:168–172. [PubMed] [Google Scholar]
- 6.Uehara Y, Hori M, Takeuchi O, Umezawa H. Mol Cell Biol. 1986;6:2198–2206. doi: 10.1128/mcb.6.6.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon H J, Yoshida M, Fukui Y, Horinouchi S, Beppu T. Cancer Res. 1992;52:6926–6930. [PubMed] [Google Scholar]
- 8.Shindo O N, Makabe O, Nagahara H, Nishimura S. Basic Life Sci. 1990;52:309–312. doi: 10.1007/978-1-4615-9561-8_26. [DOI] [PubMed] [Google Scholar]
- 9.Hoshikawa Y, Kwon H J, Yoshida M, Horinouchi S, Beppu T. Exp Cell Res. 1994;214:189–197. doi: 10.1006/excr.1994.1248. [DOI] [PubMed] [Google Scholar]
- 10.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- 11.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 12.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 13.Grunstein M. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 14.Loidl P. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- 15.Pazin M, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 16.Wolffe A P. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto M, Matsutani S, Sugita K, Yoshida H, Hayashi F, Terui Y, Nakai H, Uotani N, Kawamura Y, Matsumoto K, Shoji J, Yoshida T. J Antibiot. 1992;45:879–885. doi: 10.7164/antibiotics.45.879. [DOI] [PubMed] [Google Scholar]
- 18.Shimada J, Kwon H J, Sawamura M, Schreiber S L. Chem Biol. 1995;2:221–228. doi: 10.1016/1074-5521(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 19.Sugita K, Yoshida H, Matsumoto M, Matsutani S. Biochem Biophys Res Commun. 1992;182:379–387. doi: 10.1016/s0006-291x(05)80156-1. [DOI] [PubMed] [Google Scholar]
- 20.Taunton J, Collins J L, Schreiber S L. J Am Chem Soc. 1996;118:10412–10422. [Google Scholar]
- 21.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 22.Panyim S, Chalkley R. Arch Biochem Biophys. 1969;130:337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- 23.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 24.Kwon H J, Yoshida M, Muroya K, Hattori S, Nishida E, Beppu T, Horinouchi S. J Biochem. 1995;118:221–228. doi: 10.1093/oxfordjournals.jbchem.a124882. [DOI] [PubMed] [Google Scholar]
- 25.Riggs M G, Whittaker R G, Neumann J R, Ingram V R. Nature (London) 1977;268:462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- 26.Shute R E, Dunlap B, Rich D J. J Med Chem. 1987;30:71–78. doi: 10.1021/jm00384a013. [DOI] [PubMed] [Google Scholar]
- 27.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 28.Durrin L K, Mann R K, Kayne P S, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 29.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 30.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 31.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka M, L, M, Ogiso Y, Fujita H, Moriya S, Fruuchi K, Harabayashi T, Shinohara N, Koyanagi T, Kuzumaki N. Cancer Res. 1995;55:3228–3232. [PubMed] [Google Scholar]
- 33.Kwon H J. Ph.D. thesis. University of Tokyo; 1995. [Google Scholar]
- 34.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darkin-Rattray S J, Gurnett A M, Myers R W, Dulski P M, Crumley K M, Allocco J J, Cannova C, Meinke P T, Colletti S L, Bednarek M A, Singh S B, Goetz M A, Dombrowski A W, Polishook J D, Schimatz D M. Proc Natl Acad Sci USA. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oikawa T, Onozawa C, Inose M, Sasaki M. Biol Pharm Bull. 1995;18:1305–1307. doi: 10.1248/bpb.18.1305. [DOI] [PubMed] [Google Scholar]
- 38.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]