Abstract
Phenylglyoxal is an arginine-specific reagent that inactivates creatine kinase (CK). Previous results suggest that modification of the dimeric enzyme at a single arginine residue per subunit causes complete inactivation accompanied by the loss of nucleotide binding; the actual site of modification was not identified. Here, high-resolution tandem mass spectrometry (MS/MS) was used to identify three phenylglyoxal-modified Arg residues in monomeric rabbit muscle CK. Electrospray ionizaton Fourier-transform MS of the phenylglyoxal-modified CK that had lost ≈80% activity identified three species: unmodified, once-modified (+116 Da), and twice-modified (+232 Da) enzyme in a ratio of approximately 1:4:1. MS/MS restricts the derivatized sites to P122-P212 and P283-V332, whereas MS of Lys-C digestions revealed two modified peptides, A266-K297 and G116-K137. The only Arg in A266-K297 is Arg-291 (invariant), whereas MS/MS of modified G116-K137 shows that two of the three sites Arg-129, Arg-131, or Arg-134 (all invariant) can contain the modification. The recently reported x-ray crystal structure for the octameric chicken mitochondrial CK indicates that its nucleotide triphosphate-binding site indeed contains the equivalent of R291, R129, and R131 reported here to be at the active site of rabbit muscle CK.
Keywords: Fourier-transform mass spectrometry, electrospray ionization, protein sequencing
Creatine kinase (CK, EC 2.7.3.2) plays a key role in energy metabolism in cells by catalyzing the reversible transfer of a phosphoryl group from phosphocreatine to MgADP to form creatine and MgATP. Mechanistic studies of CK have been made more difficult by the microheterogeneity found in enzyme preparations (1); mass spectrometry (MS) recently has shown (2) that deamidation is a major cause of this. Microheterogeneity also has been an obstacle to obtaining x-ray crystal structures for dimeric cytosolic (muscle, brain, or muscle-brain heterodimer) isoforms (1, 2), although recently Fritz-Wolf et al. (3) have succeeded in obtaining a 3.0 Å x-ray crystal structure for the octameric chicken mitochondrial CK. Several studies have been done to identify the roles of amino acid residues involved in substrate binding and catalysis in CK. The reactivity of Cys-282 is well established (4, 5), though its precise catalytic/structural role is a subject of controversy (6–8). Trp-223 in mitochondrial CK (corresponding to Trp-227 in the rabbit muscle isozyme) has been suggested as an essential Trp (9), and the crystal structure of the octameric mitochondrial isoform shows this residue to be near the adenine base of ATP (3). Recent site-directed mutagenesis and chemical modification studies suggest that His-295 may be at the active site (10); however, this residue is unlikely to be the histidine implicated as a general acid/base catalyst by kinetic studies (11). In addition, evidence suggests that lysine (12, 13) and arginine (14, 15) residues may play key roles in binding ATP, presumably through electrostatic interactions with the negatively charged triphosphate; however, the specific sites of these key residues have remained elusive.
Borders and Riordan (14) showed unequivocally that full inactivation of rabbit muscle CK is accompanied by the incorporation of one phenylglyoxal (1:1 stoichiometry) as well as loss of one arginine per subunit. The actual location of the modified arginine in the primary sequence was not determined. In the mitochondrial x-ray structure, there are seven arginine residues (at positions 91, 125, 127, 231, 287, 315, and 336 corresponding to positions 95, 129, 131, 235, 291, 319, and 340 in rabbit muscle CK) near the ATP binding site, but the resolution is not sufficient to ascertain which are directly involved in ATP binding (3). Here, a combined approach of labeling of rabbit muscle CK with phenylglyoxal and high-resolution electrospray ionization (ESI) tandem Fourier transform (FT) MS is used to locate active arginines. Recent reports have demonstrated the utility of the combined labeling and ESI-MS approach to identify active residues within enzymes (16–19) as large as 42 kDa (19). Tandem ESI–FTMS also has resolved conflicts in published sequences for human CK isoforms (20).
EXPERIMENTAL PROCEDURES
Materials and Sample Preparation.
Bacterial media were produced from materials purchased from Difco. Protein chromatography reagents were from Bio-Rad and Pharmacia Biotech. Phenylglyoxal monohydrate was obtained from Sigma, and Lys-C used in the digestions was obtained from Boehringer-Mannheim. Overexpression and purification of the rabbit muscle CK was achieved by published methods (1, 10).
Modification with Phenylglyoxal.
Rabbit muscle CK (1 mg, 43 μM subunits) was equilibrated in 50 mM Bicine, 50 mM NaHCO3, pH 8.7, before treatment with 400 μM phenylglyoxal in a final volume of 500 μl at 30°C. The rate of modification was monitored by the pH stat method (4) as the loss of activity of the rabbit muscle CK as a function of time. The average half-life under these conditions was shown to be 20 ± 2 min (average of three experiments; data not shown). At 15% residual activity, the reaction was quenched and excess phenylglyoxal removed by chromatography over a HiTrap G200 column (Pharmacia) and monitored by changes in A280. The modified enzyme (or unmodified in the control) eluted in a final volume of ≈2 ml and was concentrated in a Centricon 30 ultrafiltration device (Amicon) before re-equilibration in 50 mM Tris, pH 8.8, 0.1 mM EDTA. A control experiment was carried out simultaneously under the same conditions but in the absence of the phenylglyoxal.
Enzyme Activity Measurements.
CK activity was determined at 30°C in the direction of phosphocreatine formation by the pH-stat method (4) using a Radiometer VIT90/ABU91. For the determination of specific activity or to follow the inactivation by phenylglyoxal, each 3.0 ml assay, pH 9.0, contained 40 mM creatine, 5.0 mM MgATP, 1.0 mM excess magnesium acetate, 0.1 mM EDTA, and 0.1% BSA.
Digestion with Endoproteinase Lys-C.
After equilibration in the appropriate buffer, both the prostaglandin-modified CK and the control samples were incubated with 2.5 mg of endoproteinase Lys-C in the presence of 0.1 M urea for 8 hr at 37°C. The samples then were prepared for MS by reverse-phase chromatography over a Waters Sep-Pak column, eluting in 95:5 acetonitrile/H2O gradient. The samples then were dried to completion with a Speedvac system (Savant) and resuspended in water.
ESI-FTMS.
Phenylglyoxal-modified CK samples were prepared (except where indicated) as 20 μM solutions in methanol/water/acetic acid, 76:22:2 (by vol) and electrosprayed through a syringe needle biased at 2.6–2.9 kV (3.5 kV for pure aqueous solutions) through a heated metal capillary (≈110°C) at flow rates of 0.5–1.0 μl/min. The ESI-generated ions were transported by three radio-frequency-only quadrupoles through five stages of differential pumping to the trapped ion cell (10−9 Torr, 5-s accumulation) in a modified 6.1 Tesla Finnigan FT/MS-2000 (21). MS/MS experiments were performed by using nozzle/skimmer (NS) (22) or sustained off-resonance irradiation collisionally activated dissociation (SORI-CAD) (23, 24) of charge states isolated by stored waveform inverse FT (SWIFT) (25) under conditions previously described (2). Although the resolving power nominally was 105, in the complex product mixture the 43-kDa molecular ion isotopic peaks were not resolved after deconvolution (26, 27), so that their centroid masses were used.
RESULTS AND DISCUSSION
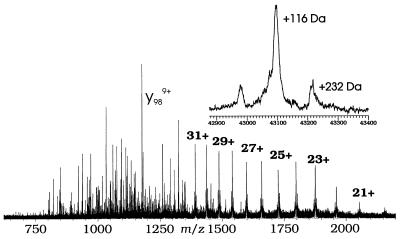
The mass spectrum of native rabbit muscle CK exhibits an isotopic cluster corresponding to a relative molecular weight (Mr) of 42,982, consistent with its DNA-derived sequence and limited deamidation (2). The spectrum of phenylglyoxal-derivatized CK (Fig. 1) shows some NS fragment ions, but is dominated by a series of (M + nH)n+ ions of molecular masses of 42,982 Da, 43,098 Da, and 43,214 Da. Phenylglyoxal has a mass of 134 Da; the observed mass shifts (+116 Da and +232 Da) are consistent with H2O loss (−18 Da) accompanying each addition (Fig. 2, discussed below). Thus, the (M + nH)n+ ions represent underivatized, once-, and twice-derivatized CK in a ratio of 1:4:1 (relative areas, Fig. 1, Inset). The modified CK used for this analysis was inactivated by phenylglyoxal to ca. 15% of the native activity. This finding suggests that the remaining unmodified CK is still fully active, whereas all the CK that has been modified is completely inactive. This result is consistent with earlier studies (14) in that any modification of CK by phenylglyoxal leads to complete inactivation. This 1:4:1 ratio shows highly selective reactivity in comparison to the other 16 arginines of rabbit muscle CK (locations are indicated in Fig. 3, Upper) (28).
Figure 1.
ESI-FTMS mass spectrum of phenylglyoxal-derivatized rabbit muscle CK. NS 140 V, sum of 25 scans. (Inset) Geometric deconvolution of all ions with masses near 43 kDa.
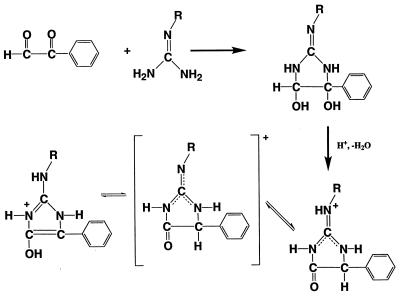
Figure 2.
Proposed mechanism for phenylglyoxal modification of reactive arginines in CK.
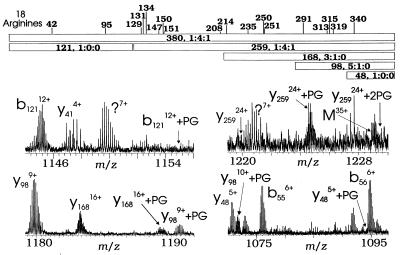
Figure 3.
(Upper) Positions of 18 arginine residues in rabbit muscle CK and (Lower) expansions of NS dissociation data from Fig. 1.
Phenylglyoxal originally was reported to modify the guanidino group of arginine with a 2:1 stoichiometry (29), and this stoichiometry is reported for most studies on this topic (30). However, inactivation of CK with phenylglyoxal is known to occur with the incorporation of only one phenylglyoxal per modified arginine (14). Although the guanidino group of arginine is very basic (pKa = 12.5 in free arginine), it is the unprotonated free base that reacts with phenylglyoxal (31). Fig. 2 shows a proposed mechanism for the formation of the +116-Da derivatized arginine, in which the unprotonated guanidino group first reacts with phenylglyoxal to form a cis-diol glyoxaline intermediate; then, under the acidic conditions used for ESI, an acid-catalyzed dehydration occurs to form the stable product.
MS/MS Site Localization.
Previous tandem ESI-FTMS of (M + nH)n+ ions from rabbit muscle CK gave 52 fragment ions whose masses can be assigned to the cDNA-derived sequence (2). Here, NS dissociation of the derivatized CK (M + nH)n+ ions was used to identify fragment ions increased in mass by 116 Da and thus modified by phenylglyoxal. The complementary b121/y259 pair represents all 380 amino acids (Fig. 3); the b121 ion shows no evidence of derivatization, whereas its complementary y259 shows unmodified, once-, and twice-derivatized CK in an approximate ratio of 1:4:1, consistent with that of the (M + nH)n+ ions**. Thus the NS dissociation evidence suggests that both sites of modification must be within the 259 C-terminal residues, eliminating Arg-42 and Arg-95. The y168 fragments (Fig. 3) are a mixture of no derivatization and a single site, with relative abundances of 3:1. Thus, one of the two derivatization sites must be in the region P122-P212, at Arg residues 129, 131, 134, 147, 150, 151, or 208. The y98 label ratio of 5:1 is similar to that that of y168, but there is little evidence for any modification of y48, localizing the second site to the region P283-V332 that contains Arg residues 291, 313, 315, and 319. However, higher-energy NS dissociation conditions did not induce fragmentation within these two identified derivatization regions, preventing further analysis of the derivatized intact protein.
Proteolysis.
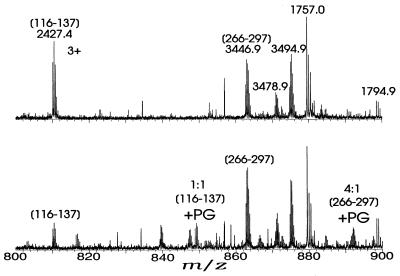
Lys-C digestion of underivatized rabbit muscle CK followed by ESI-FTMS of the unseparated mixture gave species with the masses shown in Table 1. Sixteen peptides were identified. The phenylglyoxal-derivatized CK, similarly subjected to Lys-C digestion, gave ESI-FTMS data indicating that two of the Lys-C peptides had been derivatized. The (M + 4H)4+ ion for the A266-K297 peptide (Fig. 4) shows a 4:1 ratio vs. the ion with +116 Da, indicating derivatization at Arg-291, the only arginine residue in this region. No evidence was found for derivatization in either F307-K318 or R319-K357. Arg-291 corresponds to Arg-287 in the octameric mitochondrial CK, which is one of seven arginines found at the ATP binding site (3), and is invariant (32). Interestingly, the rabbit muscle CK mutant R291Q is inactive, and the R291K mutant has approximately 1% activity (C.L.B. and G.L.K., unpublished results).
Table 1.
Peptide Mr values from Lys-C digestion of rabbit muscle CK
| Mr | Sequence (error) | Mr | Sequence (error) |
|---|---|---|---|
| 913.5 | P[1–8]K (−0.1) | 682.4 | Y[172–176]K (0.0) |
| 1,195.6 | S[15–24]K (−0.1) | 3,015.4 | P[196–222]K (−0.3) |
| 947.6 | V[32–39]K (−0.1) | 2,330.4 | S[223–241]K (−0.3) |
| 1,757.0 | D[86–100]K (−0.1) | 905.6 | I[259–265]K (−0.3) |
| 2,198.2 | D[86–104]K (−0.1) | 3,446.9 | A[266–297]K (−0.3) |
| 1,082.6 | T[107–115]K (−0.1) | 1,545.0 | F[307–316]K (−0.1) |
| 2,427.4 | Q[116–137]K (−0.1) | 3,931.3 | R[319–357]K (−0.3) |
| 1,506.9 | L[156–169]K (−0.1) | 1,301.7 | G[369–380]K (−0.1) |
Fragment masses that could not be matched against expected Mr Lys-C peptides: 1,054.8, 1,339.7, 1,528.9, 1,794.9, 2,253.3, 2,389.2, 3,478.9, 3,949.9, 3,969.2, 4,342.5, and 4,410.3 Da.
Figure 4.
Partial ESI-FTMS spectrum (m/z 800–900) for Lys-C digests of (Upper) unmodified and (Lower) prostaglandin-derivatized CK.
The G116-K137 peptide also shows a +116-Da mass shift at reaction with phenylglyoxal, with a 1:1 ratio of the underivatized and once-derivatized for the 3+ charge state (Fig. 4) and 1:2 for the 4+ charge state. This shift restricts the second reactive site to Arg-129, Arg-131, or Arg-134, all of which are invariant in all CK isoforms (32). The 3+ ions of the once-derivatized G116-K137 peptide were accumulated (33, 34) in the FTMS trap for 10 cycles by using selective stored waveform inverse FT (SWIFT) (25) waveforms and quadrupolar axialization (33–36), but sustained off-resonance irradiation-collisionally activated dissociation (SORI-CAD) (−1.1 kHz, ≈3 Vpp) of these (22, 23) gave only P122-I136 as the smallest fragment ion containing the phenylglyoxal; higher collision energies and infrared multiphoton dissociation (37) produced no other identifiable fragments. Thus it was not possible to delineate among Arg-129, Arg-131, or Arg-134 as the site of phenylglyoxal derivatization. However, both the 3+ (Fig. 4) and 4+ charge states of the derivatized Lys–C peptides show a doubly labeled G116–K137 peptide in a concentration that is ≈20% of that of the singly labeled peptide, indicating that two of these arginines are accessible to some extent. However, consistent with this finding, the octameric mitochondrial CK structure indicates that its Arg-125 and Arg-127 (corresponding to Arg-129 and Arg-131, respectively, in rabbit muscle CK) are at the ATP binding site (3).
CONCLUSIONS
The combined approach of chemical modification, Lys-C digestion, and tandem ESI-FTMS has revealed that phenylglyoxal inactivation takes place at three sites, at Arg-291 and partially at two of the sites Arg-129, Arg-131, or Arg-134; a recent x-ray crystal structure shows the equivalent of Arg-291, Arg-129, and Arg-131 to be at the active site. This approach also could identify residues in other ATP-binding enzymes for which no x-ray structure has been solved, such as glucokinase (38).
Acknowledgments
We especially thank Neil L. Kelleher, Peter B. O’Connor, Michael W. Senko, Patricia C. Babbitt, Camille B. White, Einar Fridriksson, Gary A. Valaskovic, Sajid Bashir, and Craig P. Dufresne for helpful comments and suggestions during the course of this work. This work was supported by National Institutes of Health Grants GM 16609 (to F.W.M.) and AR17323 (to G.L.K.), Research Corporation Grant CC4075 (to C.L.B.), the State University of New York at Buffalo (to T.D.W.), and American Heart Association Postdoctoral Fellowship 91–06 (to L.H.C.).
ABBREVIATIONS
- CK
creatine kinase
- ESI
electrospray ionization
- MS
mass spectrometry
- FT
Fourier transform
- MS/MS
tandem mass spectrometry
- NS
nozzle/skimmer dissociation
Footnotes
The relative abundances of different charge states of the same ion are a function of the ion’s structure, so that the ratio of an ion’s abundance to that of its derivatized counterpart can vary with charge value.
References
- 1.Chen L H, Babbitt P C, Vásquez J R, West B L, Kenyon G L. J Biol Chem. 1991;266:12053–12057. [PubMed] [Google Scholar]
- 2.Wood T D, Chen L H, Kelleher N L, Little D P, Kenyon G L, McLafferty F W. Biochemistry. 1995;34:16251–16254. doi: 10.1021/bi00050a004. [DOI] [PubMed] [Google Scholar]
- 3.Fritz-Wolf K, Schnyder T, Wallimann T, Kabsch W. Nature (London) 1996;381:341–345. doi: 10.1038/381341a0. [DOI] [PubMed] [Google Scholar]
- 4.Mahowald T A, Noltmann E A, Kuby S A. J Biol Chem. 1962;237:1535–1548. [PubMed] [Google Scholar]
- 5.Buechter D D, Medzihradszky K F, Burlingame A L, Kenyon G L. J Biol Chem. 1992;267:2173–2178. [PubMed] [Google Scholar]
- 6.Kenyon G L, Reed G H. Adv Enzymol. 1983;54:367–426. doi: 10.1002/9780470122990.ch6. [DOI] [PubMed] [Google Scholar]
- 7.Hou L X, Vollmer S. Biochim Biophys Acta. 1994;1205:83–88. doi: 10.1016/0167-4838(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 8.Fuerter R, Furter-Graves E M, Walliman T. Biochemistry. 1993;32:7022–7029. doi: 10.1021/bi00078a030. [DOI] [PubMed] [Google Scholar]
- 9.Gross M, Furter-Graves E M. Protein Sci. 1994;3:1058–1068. doi: 10.1002/pro.5560030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L H, Borders C L, Jr, Vásquez J R, Kenyon G L. Biochemistry. 1996;35:7895–7902. doi: 10.1021/bi952798i. [DOI] [PubMed] [Google Scholar]
- 11.Cook P F, Kenyon G L, Cleland W W. Biochemistry. 1981;20:1204–1210. doi: 10.1021/bi00508a023. [DOI] [PubMed] [Google Scholar]
- 12.Kassab R, Roustan C, Pradel L. Biochim Biophys Acta. 1968;167:308–316. doi: 10.1016/0005-2744(68)90210-6. [DOI] [PubMed] [Google Scholar]
- 13.James T L, Cohn M. J Biol Chem. 1974;249:2599–2604. [PubMed] [Google Scholar]
- 14.Borders C L, Jr, Riordan J F. Biochemistry. 1975;21:4699–4704. doi: 10.1021/bi00692a021. [DOI] [PubMed] [Google Scholar]
- 15.James T L. Biochemistry. 1976;15:4724–4730. doi: 10.1021/bi00666a029. [DOI] [PubMed] [Google Scholar]
- 16.Aplin, R. T., Robinson, C. V., Schofield, C. J. & Waley, S. G. (1993) J. Chem. Soc. Chem. Commun. 121–123.
- 17.Aplin R T, Robinson C V, Schofield C J, Westwood N J. Tetrahedron. 1993;49:10903–10912. [Google Scholar]
- 18.Roach, P. L., Baldwin, J. E., Aplin, R. T., Robinson, C. V. & Schofield, C. J. (1994) J. Chem. Soc. Chem. Commun. 849–850.
- 19.Kelleher N L, Costello C A, Begley T P, McLafferty F W. J Am Soc Mass Spectrom. 1995;6:981–984. doi: 10.1016/1044-0305(95)00584-Z. [DOI] [PubMed] [Google Scholar]
- 20.Wood T D, Chen L H, White C B, Babbitt P C, Kenyon G L, McLafferty F W. Proc Natl Acad Sci USA. 1995;92:11451–11455. doi: 10.1073/pnas.92.25.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beu S C, Senko M W, Quinn J P, Wampler F M, III, McLafferty F W. J Am Soc Mass Spectrom. 1993;4:557–565. doi: 10.1016/1044-0305(93)85017-R. [DOI] [PubMed] [Google Scholar]
- 22.Loo J A, Edmonds C G, Smith R D. Anal Chem. 1991;63:2488–2499. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 23.Gauthier J W, Trautman T R, Jacobson D B. Anal Chim Acta. 1991;246:211–225. [Google Scholar]
- 24.Senko M W, Speir J P, McLafferty F W. Anal Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 25.Marshall A G, Wang T-C L, Ricca T L. J Am Chem Soc. 1985;107:7893–7897. [Google Scholar]
- 26.Mann M, Meng C K, Fenn J B. Anal Chem. 1989;61:1702–1708. [Google Scholar]
- 27.O’Connor P B, McLafferty F W. J Am Chem Soc. 1995;117:12826–12831. [Google Scholar]
- 28.Putney S, Herlihy W, Royal N, Pang H, Aposhian H V, Pickering L, Belagaje R, Biemann K, Page D, Kuby S, Schimmel P. J Biol Chem. 1984;259:14317–14320. [PubMed] [Google Scholar]
- 29.Takahashi K. J Biol Chem. 1968;243:6171–6179. [PubMed] [Google Scholar]
- 30.Riordan J F. Mol Cell Biochem. 1979;26:71–92. doi: 10.1007/BF00232886. [DOI] [PubMed] [Google Scholar]
- 31.Bjerrum P J, Wieth J O, Borders C L., Jr J Gen Physiol. 1983;81:453–484. doi: 10.1085/jgp.81.4.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mühlebach S M, Gross M, Wirz T, Wallimann T, Perriard J-C, Wyss M. Mol Cell Biochem. 1994;133/134:245–262. doi: 10.1007/BF01267958. [DOI] [PubMed] [Google Scholar]
- 33.Bruce J E, Anderson G A, Hofstadler S A, Van Orden S L, Sherman M S, Rockwood A L, Smith R D. Rapid Commun Mass Spectrom. 1993;7:914–919. [Google Scholar]
- 34.Hasse H U, Becker S, Dietrich G, Klisch N, Kluge H J, Lindinger M, Lutzendirchen K, Schweikhard L, Ziegler J. Int J Mass Spectrom Ion Processes. 1994;132:181–191. [Google Scholar]
- 35.Speir J P, Gorman G S, Pitsenberger C C, Turner C A, Wang P P, Amster I J. Anal Chem. 1993;65:1746–1752. doi: 10.1021/ac00061a018. [DOI] [PubMed] [Google Scholar]
- 36.Guan S, Kim H S, Marshall A G, Wahl M C, Wood T D, Xiang X. Chem Rev. 1994;94:2161–2182. [Google Scholar]
- 37.Little D P, Speir J P, Senko M W, O’Connor P B, McLafferty F W. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 38.Pilkis S J, Weber I T, Harrison R W, Bell G I. J Biol Chem. 1994;269:21925–21928. [PubMed] [Google Scholar]