Abstract
The protection against micrococcal nuclease digestion afforded to chromatosomal DNA by the presence of a linker histone (H1o) has been quantitatively measured in two reconstituted systems. We have used chromatosomes reconstituted at two distinct positions on a DNA fragment containing the 5S rRNA gene from Lytechinus variegatus and at a specific position on a sequence containing Gal4- and USF-binding sites. In all cases, we find asymmetric protection, with ≈20 bp protected on one side of the core particle and no protection on the other. We demonstrated through crosslinking experiments that the result is not due to any sliding of the histone core caused by either linker histone addition or micrococcal nuclease cleavage. Because the core particle is itself a symmetric object, the preferred asymmetric location of a linker histone must be dictated by unknown elements in the DNA sequence.
Lysine-rich histones (H1, H5, and their cognates) bind to linker DNA in the chromatin fiber (hence their name, linker histones, LH) and contribute to the formation/maintenance of higher-order fiber structures (for a recent review, see ref. 1). The discovery of the chromatosome—the nucleosomal particle containing about 168 bp of DNA wrapped around the histone octamer and one molecule of linker histone (2)—prompted research into understanding how the linker histone binds to the nucleosome particle. Despite two decades of effort, the issue is still highly contentious. Earlier data suggested that the LH or its isolated globular domain (GD) bound near the dyad axis of the particle, with 10 bp of DNA protected against micrococcal nuclease (MNase) on each side of the core particle (2–5). This view is consistent with evidence for two DNA-binding sites on opposite sides of the GD (6), both of which are required for the formation of the chromatosome (7).
More recently, alternative, asymmetric placements of the GD have been proposed. Wolffe, Hayes, and collaborators (8–10) studied the location of either intact LHs or their isolated GDs on chromatosomes reconstituted on 5S rRNA genes from Xenopus borealis. On this sequence, they reported that LH protects linker DNA asymmetrically, 5 bp on one side, and 15 bp on the other. DNA–protein crosslinking experiments further demonstrated that the GD of histone H5 contacted DNA at a site 65 bp away from the dyad axis, on only one side of the particle (9). More recent experimental results from the same authors placed the GD within the DNA gyres of the nucleosomal particle (11, 12). An alternative off-axis model has been proposed by Travers and Muyldermans (13) on the basis of careful statistical analysis of DNA sequence elements in chromatosome DNA (13, 14). This model postulates that the GD lies on the outside of the particle, bridging two adjacent DNA gyres. The actual contacts are proposed to be with DNA at positions ±7 and ±1 of the DNA superhelix. For an excellent critical evaluation of these models of possible LH locations, see Crane-Robinson (15). Thus, although the conformation of the nucleosomal core particle is now known in exquisite detail (16–18), the structure of the next higher element of the chromatin fiber remains in debate.
In an attempt to better understand how the GD in the Wolffe/Hayes model can protect DNA on both sides of the core particle, albeit asymmetrically, we have recently repeated the MNase protection experiments by using chromatosomes reconstituted on the X. borealis 5S sequence. The reconstitution proceeded in two stages. First, a histone octamer was added to the DNA to produce what we will call a core nucleosome. Then one molecule of LH was added to make a chromatosome. We found that, with this sequence, results interpreted as “protection” by LH can be obtained with either naked DNA or with reconstituted core nucleosomes, in the absence of linker histones (W.A., K.v.H., and J.Z., unpublished data). This means that the X. borealis sequence is not a reliable matrix for such studies. Therefore, we have sought alternative positioning sequences to determine the pattern of protection of linker DNA by linker histones.
The 5S rDNA from the sea urchin Lytechinus variegatus was an obvious choice, because the positioning of the core particle is well understood on this sequence (19–21). In addition, this is the only sequence, apart from that of X. borealis 5S rDNA, on which chromatosome positioning has been studied (22). These latter experiments, however, made use of tandemly repeated 5S rRNA genes to allow the formation of short chromatin fibers, and it is not clear, on an a priori basis, whether the chromatosome positions on fibers will be the same as those on individual particles. The other sequence we used is totally unrelated to the 5S gene sequences. It contains GAL4- and USF-binding sites on a DNA fragment that was reported to provide two major binding sites for the histone octamer (23). Our results show that the LH provides protection of linker DNA on one side of the core particle only. The choice by LH of which side to protect, and hence where to bind in the core nucleosome, depends on the DNA sequence used in reconstitution.
MATERIALS AND METHODS
Expression of Human H1°.
Human H1° gene was obtained by PCR of pWH312 (24) and cloned into pET-15b expression vector, as recently detailed (W.A. et al., unpublished data).
DNA Fragments Used for Reconstitution.
To improve the yield of DNA for reconstitution, DNA fragments were recloned as tandem repeats. The 179-bp fragment obtained by BamHI digestion of pGUB (ref. 23, plasmid kindly provided by J. Workman, Pennsylvania State University) was recloned into the BamHI site of plasmid pUC19. Digestion of the resulting plasmid with BamHI produced the original 179-bp fragment, and digestion with EcoRI and HindIII produced a 235-bp fragment containing 30- and 20-bp extensions on the 5′ and 3′ sides of the 179-bp fragment, respectively (Fig. 1A). The 243-bp BamHI fragment containing the sea urchin 5S rRNA gene was obtained by cloning a 195-bp EcoRI fragment from p5S207–12 (ref. 25, plasmid courtesy of R. Simpson, Pennsylvania State University) into the EcoRI site of pBSIISK+, obtaining a 243-bp copy of this sequence by extending it on both sides by ≈20 bp by PCR while introducing BamHI sides on the ends, and finally cloning it into the BamHI site of pUC19 as a tandem repeat of four copies. The sequence of the resulting BamHI-BamHI fragment is presented in Fig. 1A. The recloned fragments were obtained after digestion with the respective restriction endonucleases and purified from agarose electrophoresis by electroelution (Schleicher & Schuell).
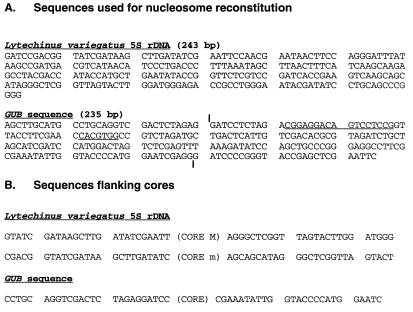
Figure 1.
(A) Nucleotide sequences of DNA fragments used for reconstitution. The vertical bars in the GUB sequence denote the ends of the shorter 179-bp fragment also used for reconstitution; the sequences constituting GAL4- and USF-binding sites (23) are underlined. (B) Sequences flanking the core particles identified in this study. Cores M and m are as defined in Fig. 3B.
Reconstitution of Core Nucleosomes and Chromatosomes.
Reconstitutions were carried out by the salt dialysis methods of Tatchell and van Holde (26) at 4°C. Core histone octamers (27) were mixed with the respective DNA fragments at a molar ratio of ≈0.6 octamers/DNA to obtain nucleosome occupancy on ≈50% of DNA fragments (8), and then dialyzed successively vs. decreasing concentrations of NaCl down to 0.05 M NaCl. H1° was mixed with reconstituted core particles in 50 mM NaCl and TE (10 mM Tris⋅HCl, pH 7.5/0.25 mM EDTA) and incubated at 23°C for 30 min. The success of reconstitution was monitored by electrophoresis in 0.9% agarose gels in 0.5× TBE (0.045 M Tris⋅borate, pH 8.0/1 mM EDTA). The gels were stained with 1 μg of ethidium bromide (EtdBr) per ml and then destained in the electrophoresis buffer for 1 h before photography.
MNase Digestion of Reconstituted Core Nucleosomes and H1°-Containing Chromatosomes.
The solution containing reconstituted nucleosomes was made 1 mM with respect to CaCl2, and 2 units of MNase (Worthington) were added per 5 μg of DNA. Digestion was stopped after 5 min by bringing the solution to 6 mM EDTA/0.4% SDS and placing the tube on ice for 10 min. One hundred micrograms of Proteinase K per ml was then added, and the sample was incubated for 1 h at 37°C. DNA was twice phenol-extracted and ethanol-precipitated, and the pellet was dissolved in 10 μl of 10 mM Tris⋅HCl, pH 7.5/0.25 mM EDTA.
Gel Purification of Chromatosome and Core Particle DNA, End-labeling, and Restriction Nuclease Digestion.
DNA from MNase digests was electrophoresed in 10%, 15-cm-long polyacrylamide slab gels (Idea Scientific) in TBE (0.09 M Tris⋅borate, pH 8.0/2 mM EDTA) at 13 V/cm for 5 h. After EtdBr staining, the chromatosome and core particle DNA bands were cut out of the gel, and the gel was crushed, mixed with ≈400 μl of elution buffer (0.5 M ammonium acetate, pH 8.0/10 mM magnesium acetate/1 mM EDTA/0.1% SDS), incubated overnight in a 37°C shaker, and centrifuged for 1 min. The supernatant was passed through siliconized glass wool, phenol-extracted, ethanol-precipitated, and dissolved in 5 μl of 10 mM Tris⋅HCl, pH 8.0/1 mM EDTA. Purified DNA fragments were 5′ end-labeled and digested with restriction endonucleases according to standard procedures. The products of digestion were analyzed either on 15% nondenaturing polyacrylamide gels run in TBE or on 6% denaturing sequencing gels run under standard conditions (28).
Crosslinking of Histone Octamer to DNA in Reconstituted Core Nucleosomes.
Reconstituted core nucleosomes dialyzed to TE (10 mM Tris⋅HCl, pH 8.0/0.25 mM EDTA) were treated with glutaraldehyde (10% electron microscopy-grade solution from EM Science) to 0.1% final concentration. Samples were incubated at 4°C for 8 h with gentle shaking and dialyzed overnight vs. TE containing 50 mM NaCl. The success of crosslinking was determined on SDS-polyacrylamide gels (29) and 0.9% agarose gels.
RESULTS
Experimental Approach.
Fig. 1A presents the DNA sequences used to study the positions of the core particles and chromatosomes. The DNA fragments of interest were reconstituted with histone octamers, followed by the addition of LH according to published procedures (see Materials and Methods). Reconstitution was monitored by band shift analysis like that illustrated in Figs. 2A and 4A. The position of the reconstituted core particles and chromatosomes on the DNA fragments was determined by the method originally introduced by Dong et al. (20). In this approach, the reconstituted particles were subjected to MNase digestion to trim down the unprotected linker DNA, and the digested DNA was purified and fractionated on DNA electrophoretic gels. DNA bands of defined lengths (145 bp representing the core particle and 168 bp representing the chromatosome, but see also below) were eluted from the gels and subjected to restriction nuclease digestion, and the lengths of the resulting DNA fragments were determined by electrophoresis on polyacrylamide sequencing gels. Mild micrococcal nuclease digestion was also carried out on naked DNAs for controls.
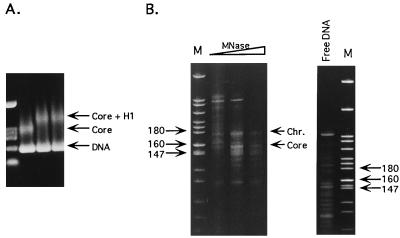
Figure 2.
Chromatosome reconstitution on the 5S rDNA fragment from L. variegatus. (A) Reconstitution of DNA into core nucleosomes and chromatosomes as visualized by the band shift assay. The 243-bp fragment was reconstituted with core histone octamer and further with histone H1° (LH/core histone molar ratios of 1.0 and 1.3 in lanes 3 and 4, respectively). The resultant complexes were resolved in 0.9% agarose gels (see Materials and Methods). Lane 1 contains pBSIISK+/HinfI, XbaI marker. (B) Products of MNase digestion of the mixture of chromatosomes and naked DNA, presented in A, lane 4. Lanes labeled M contain pBR322/MspI size markers. The open triangle above the lanes indicates increasing levels of MNase digestion. The positions of core- and chromatosome-sized DNA fragments are indicated. The control pattern of digestion of free DNA is also presented. The free DNA present in the incubation mixture will not contribute to the patterns observed with the core or chromatosome reconstitutes, because under the conditions of digestion used to obtain DNA from these particles naked DNA is completely digested to small fragments unobservable on gels.
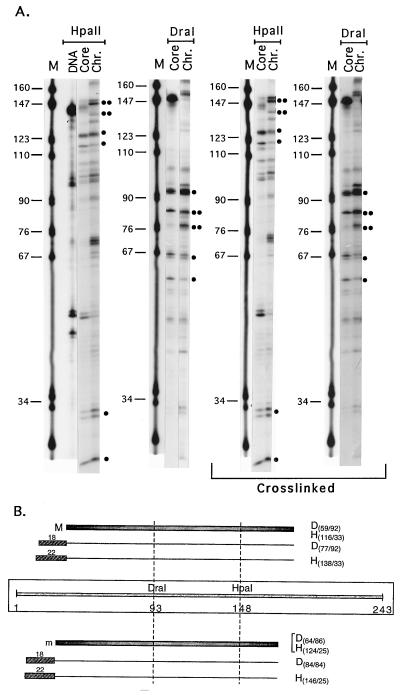
Figure 4.
Chromatosome mapping on the GUB sequence. (A) Reconstitution of DNA into core particles and chromatosomes as visualized by the band shift assay. Lanes: 1, the pBSIISK+/HinfI, XbaI marker fragments; 2, naked DNA; 3, core nucleosome reconstitution; 4, chromatosome reconstitution (LH/core histone molar ratio of 1.3). (B) Products of MNase digestion of the mixture of chromatosomes and naked DNA, presented in A, lane 4. The positions of the chromatosome- and core-nucleosome DNA are marked with arrows on the left-hand side of the gel; the arrows on the right side denote the positions of the marker fragments of interest. The open triangle above the lanes indicates increasing levels of MNase digestion. Lane denoted free DNA shows the products of MNase digestion of naked DNA. Note the absence of any fragments of core or chromatosome lengths. (C) Linker histone-induced protection of linker DNA against MNase digestion. Lanes: M, pBR322/MspI size markers; Core and Chr., restriction fragments of core- and chromatosome-sized DNA fragments extracted from the MNase gels, such as those shown in B. Main digestion products are marked by dots, as in Fig. 3. (D) Scheme illustrating the protection results. For further details see legend to Fig. 3B.
The electrophoretic patterns on such gels were usually rather complex because of the existence of alternative positions of the particles along the sequence and, even more so, because strong, sequence-dependent sites for MNase cleavage would be cleaved even in the presence of histones. This complexity required extreme care in the interpretation of the results. First, the experiments were repeated at least three times on independent preparations of reconstituted particles for each DNA fragment used. Second, DNA fragments that were present in both particle digests and in digests of the corresponding naked DNA were not considered to reliably reflect nucleosome positions. From the remaining fragments, the only pairs taken in account were those whose length would sum up to the length of DNA expected for either the core or the chromatosome particles. Finally, the results obtained with one restriction enzyme were compared with those obtained with another enzyme.
Linker Histone Protection on Chromatosome Particles Reconstituted on 5S rDNA from L. variegatus.
The band shift analysis used to monitor the success of nucleosome particle reconstitution on the L. variegatus sequence is shown in Fig. 2A, and the pattern of DNA fragments resulting from MNase digestion of reconstituted chromatosomes is displayed in Fig. 2B. As expected, the presence of LH created the kinetic pause in the digestion pattern at ≈170 bp, characteristic of the chromatosome. It should be noted that in this case the DNA fragments corresponding to both the core particle and the chromatosome appeared to be slightly longer than the expected 146 and 168 bp when analyzed using the pBR322/MspI set of molecular markers, the core particle DNA appearing to be closer to 153 bp and that of the chromatosome ≈175 bp. This apparent discrepancy was noted before (20) and attributed to the probable existence of a slight curvature in this sequence that increases its apparent length on polyacrylamide gels.
Analysis of core particle positions on the L. variegatus sequence revealed, in accordance with earlier studies (20, 21), one major M and some minor positions (Fig. 3A, lanes marked Core), of which position m was the most prominent (Fig. 3B). Moreover, these positions were the same whether analysis was performed on DNA fragments of core particle length isolated from MNase digests of either core nucleosome or chromatosome reconstitutes, again in conformity with previous reports (22). The pattern of restriction nuclease digestion of chromatosome-length DNA fragments obtained upon MNase digestion of reconstituted chromatosomes (Fig. 3A, lanes marked Chr.) revealed a number of fragments. Those major fragments that were observed reproducibly in independent reconstitution experiments, whose length added up to chromatosome-length DNA, and which were absent in patterns obtained from naked DNA were as follows: digestion with DraI produced pairs of 77/92 and 84/84 bp fragments and digestion with HpaII produced pairs of 138/33 and 146/25 bp.
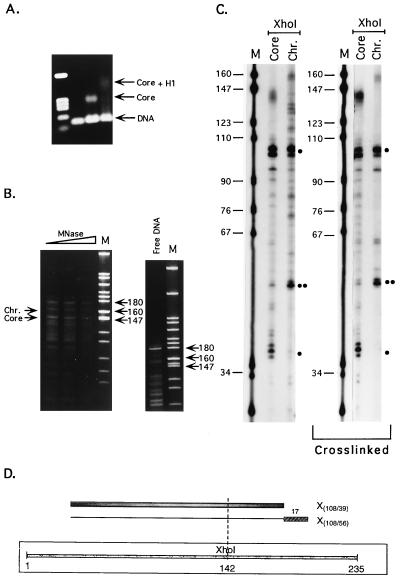
Figure 3.
(A) Linker histone-induced protection of linker DNA against MNase digestion. Lanes: M, pBR322/MspI size markers; Core and Chr., restriction fragments of core- and chromatosome-sized DNA fragments extracted from the MNase gels such as those shown in Fig. 2B. DNA designates lane containing digestion products of the core-sized DNA fragment extracted from MNase digestion gels of naked DNA. Main digestion products identified as representing core or chromatosome positions (see text) are marked by dots. One dot designates fragments seen in either the core nucleosome or in both the core and the chromatosome; two dots designate new fragments observed in chromatosome digests only. The restriction enzymes used are denoted above the respective lanes. Lanes marked crosslinked present the results of the same analysis performed after crosslinking the protein to the DNA at the core nucleosome reconstitution step, before the addition of LH. (B) Scheme summarizing the protection data. The positions of the core particles M and m are denoted as gray bars, and those of the chromatosomes are denoted as plain lines with stippled gray boxes at the end, representing the DNA stretches protected by LH binding. The numbers above these boxes denote the lengths of the protected regions (in bp). The light gray bar in the middle of the scheme represents the DNA fragment used for reconstitution, with the sites for DraI and HpaII cleavage marked accordingly. The pairs of restriction fragments used for the assignments are marked with D (for DraI) or H (for HpaII), followed by numbers denoting the lengths of the fragments, as determined from the sequencing gels.
One possible interpretation of these results in terms of linker DNA protection in the chromatosome is presented in Fig. 3B. This interpretation makes two assumptions that must be carefully evaluated. The first is that LH binding does not cause short-range sliding of the histone octamer along the DNA fragment. This question was experimentally resolved (see below). The second assumption is that each pair of chromatosome fragments results from protection of one of the two possible core particle positions. This assumption is necessitated by the absence of information about the precursor–product relationships among the positions of the cores and chromatosomes observed in the patterns of the chromatosome reconstitutes. In practical terms, it means that each chromatosome is taken to derive from the core nucleosome with which it shares one common border (see Fig. 3B).
We are confident that this second assumption is correct, because fragments D84/84 and H146/25 can only be interpreted as deriving from LH protection of core particle m if short-range shifting of the position of the histone core by LH binding is excluded (see below). In this case, the protection is only on one side of the core particle. If fragments H(138/33) and D(77/92) are assigned to core particle M, then the protection provided by LH binding is also only on one side of the core particle. However, if these same fragments are considered as derived from LH protection to core particle m, then one should interpret them as protection of 15 bp to the 5′ side and 6 bp to the 3′ side of particle m. An asymmetric, one-sided protection and a two-sided protection conferred by LHs would imply different locations of LH in the particle, off-axis and over-the-axis, respectively. Although such a situation cannot be formally ruled out on the basis of the present results, it seems highly unlikely, because if restriction fragments H(138/33) and D(77/92) are assigned to m, there will be no chromatosome corresponding to the major position M. When the data on pGUB (see below) also are taken into account, a LH protection of DNA on both sides of the core particle seems even more unlikely.
Binding of Linker Histone Does Not Shift the Position of the Core on the L. variegatus 5S rDNA.
As stated above, the interpretation of the complex restriction nuclease digestion patterns was based on the assumption that binding of LH to the core nucleosome did not cause short-range shifts of the histone octamer along the DNA fragment. Without such an assumption, each individual chromatosome position could reflect many different ways of linker DNA protection, depending on the direction and magnitude of the shift. Thus, for example, D(77/92) could reflect the protection illustrated on Fig. 3B, but the same DNA fragments could be obtained if the LH shifted the position of the M core ≈20 bp in the 5′ direction and protected ≈20 bp of (now linker) DNA at the 3′ end. To exclude such interpretations, the whole analysis was repeated after crosslinking the protein to the DNA at the core nucleosome reconstitution step, before addition of LH. The results were the same as those obtained without crosslinking (Fig. 3A, lanes marked crosslinked), reinforcing the interpretations of LH protection shown in Fig. 3B.
Linker Histone Protection on Chromatosome Particles Reconstituted on the Positioning Fragment from pGUB.
The same type of experiments were performed on the 179-bp BamHI-BamHI fragment derived from pGUB (see Materials and Methods and Fig. 1A). Because the only major position of the core particle on this fragment was only 2 bp away from its 5′ end (this work, and ref. 23), and because proper LH binding may require longer DNA stretches on both sides of the core particle, the same experiments were repeated with another fragment of the same sequence, containing an additional 30 bp at the 5′ end and 26 bp at the 3′ end (see Materials and Methods, and Fig. 1A). This longer fragment had an additional minor core position, starting at bp 11 from the 5′ end. Both fragments gave the same results with respect to LH protection of the major core position, that is, protection of ≈20 bp at the 3′ end.
Fig. 4A presents an example of the band shift experiments to monitor reconstitution, Fig. 4B shows representative gels of MNase cleavage of reconstituted chromatosomes, and Fig. 4C shows a sequencing gel of the DNA fragments obtained after restriction digestion of core- or chromatosome-length DNA extracted from gels like those in Fig. 4B. The interpretation of the results is schematically shown in Fig. 4D. It is clear that H1o protects ≈17 bp of linker DNA on only one side of the core particle. Repeating the experiment with histone octamers crosslinked to DNA before the addition of LH to preclude shifting of the core upon LH binding gave indistinguishable results (Fig. 4C). Again, as in the case of L. variegatus sequence, LH binding did not cause short-range sliding of the histone octamer along the sequence.
Thus, in the case of this sequence, LH always protects the linker DNA on one side only of the core particle, which implies that it always binds in a strictly defined asymmetric position to the core particle.
DISCUSSION
As stated in the Introduction, the issue of where LH is situated in the nucleosome is a matter of contention. In an initial attempt at resolving the existing controversies, we have recently shown that the 5S rDNA from X. borealis, widely used as a reconstitution substrate to study LH location in the nucleosomal particle, contains very strong MNase cleavage sites that may create ambiguities in interpreting the “protection” data. Therefore, in this report, we have turned to alternative DNA sequences to reconstitute core nucleosomes and chromatosomes and studied the protection created by LH binding on the “linker” DNA by established methods.
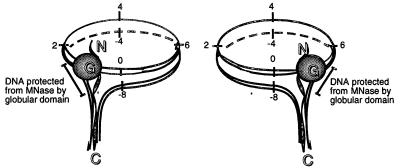
We find that (i) LH protects DNA on only one side of the core particles and that (ii) LH binding does not cause short-range sliding of the histone octamer along the DNA template. Moreover, and more importantly, we show that the asymmetric, one-sided protection of linker DNA is sequence-dependent. On some sequences, as in the positioning sequence from the L. variegatus 5S rDNA, the LH protects linker DNA on the 5′ side of the particle (5′ being defined as the 5′ end of the upper strand sequence). On other positioning sequences, like the one derived from pGUB, the protection is to the 3′ side of the core particle, if one defines the directionality of the underlying DNA sequence as the 5′ to 3′ direction of the upper DNA strand of the GAL4- and USF-binding sites (see also below for further discussion). A schematic presentation of how the LH binds to the core particle and protects DNA from MNase digestion is given in Fig. 5.
Figure 5.
Model for positioning of a molecule of linker histone on the core particle. The globular domain is presented by a solid circle, and the N- and C-terminal tails are presented as unstructured stretches. The portions of linker DNA protected by LH binding are marked. The numbers denote the number of helical turns of the DNA, with 0 denoting the dyad axis. Two alternative off-axis locations of the LH are depicted. The C-terminal tail of the LH is drawn as simultaneously interacting with the entering and exiting DNA helices to form the stem structure recently described by Hamiche et al. (32).
The model we derived from these results strongly resembles that proposed recently by Travers and Muyldermans (13) on the basis of analysis of the statistical distribution of certain di-, tri-, and tetranucleotides along the DNA in a library of chromatosomal DNA clones. These authors find that a short DNA sequence, AGGR (where r = A or G), is preferentially located at one, but not both, of the termini of the cloned DNA, making the sequence organization of chromatosomal DNA asymmetric with respect to the midpoint. By combining the sequence data with the structural distortion seen by Pehrson (5) in chromatosomal DNA at a position close to the dyad axis, they place one contact of GH5 close to the dyad; the other contact is with an adjacent gyre close to one extremity. Our data additionally show that the LH may actually choose between the two symmetrically located potential binding sites and do so in a sequence-dependent manner.
We looked in the core-flanking regions of the sequences studied here (Fig. 1B) to search for DNA sequence elements reported by Travers and Muyldermans (13) and Muyldermans and Travers (14) to flank the core particles in chromatosomal DNA. Although such elements could be identified, no general conclusion could be derived from the small sample available in this work. To unequivocally determine the contribution of specific oligonucleotide signals in linker DNA to LH positioning, engineered sequences containing such presumptive signals on either side of a positioned core particle should be analyzed.
How can earlier data indicating symmetric protection on both sides be reconciled with the new view that the chromatosome is an extension of the core particle on only one of its sides? There may be particular arguments, depending on the system and method used in earlier studies, but a major common feature of those experiments emerges: all used heterogeneous populations of nucleosome particles derived from tissues (chicken erythrocytes, refs. 2 and 4; rat liver, ref. 5). Using such particles will produce a population-averaged picture that can reflect equally well either a symmetric over-the-dyad location of the LH or approximately equal occupancy of two equivalent and slightly off-axis sites, as discussed by Crane-Robinson (15). We would like to also point out that analysis of the actual data of Simpson (2) does not allow an unequivocal conclusion to be reached. As stated by the author himself, “these results suggest that the structure of the chromatosome may involve extension of the core particle DNA by addition of 10 bp more DNA to each end” (emphasis added).
Insofar as in vivo significance is concerned, it may be relevant to mention results from Mirzabekov’s laboratory [Bavykin et al. (30)] obtained from MNase digestion of native chromatin fibers. When unfolded fibers were analyzed, the series of mononucleosome particles obtained was: MN145, MN165, MN175, MN185, etc., with the conspicuous absence of a particle containing DNA 155 bp in length (such a particle was, however, observed upon digestion of condensed fibers, for reasons that remain to be understood). The absence of the MN155 in digests of the unfolded fibers may be the result of the asymmetric protection of a longer DNA fragment (≈20 bp) on only one side of the core, like the one we have observed here on defined sequence-reconstituted nucleosomes.
That the LHs protect DNA on only one side of the core particle seems beyond reasonable doubt. In the two cases studied here, as well as in another example (W.A. et al., unpublished data), we see protection on either the 5′ or the 3′ side of the core particle (assigning arbitrary directionality of the DNA sequence), but never on both sides. Asymmetric ≈20-bp protection has also been reported very recently in another instance (31). Even if many more sequences are studied at the level of the mononucleosomal particle, and the directionality is more precisely defined (using, for instance, the direction of transcription of the transcribed strand as a criterion for assigning directionality), it would still be impossible to understand whether in the fiber the LH are always situated on one and the same side of successive core particles, or are randomly located on either one side. The answer to this question would be of immense importance to our understanding of chromatin fiber structure and would require investigation of several successive nucleosomal particles in native or reconstituted fibers.
Finally, if the inside location of the GD proposed by Pruss et al. (11) and Hayes (12) were a general property of the generic chromatosome, it would be difficult to see how it could protect ≈20 bp of linker DNA. It is obvious that additional experiments that rely on techniques complementary to the one used here should be applied to resolve the remaining puzzle of the outside vs. inside location of the linker histone in the chromatosome.
Acknowledgments
We are indebted to Drs. D. Doenecke, J. Workman, and R. Simpson for providing plasmids pWH312, pGUB, and p5S207-12, respectively. This research was supported by National Institutes of Health Grant GM50276 to K.v.H. and J.Z.
ABBREVIATIONS
- GD
globular domain
- EtdBr
ethidium bromide
- LH
linker histone
- MNase
micrococcal nuclease
References
- 1.Zlatanova J, van Holde K. Prog Nucleic Acids Res Mol Biol. 1996;52:217–259. doi: 10.1016/s0079-6603(08)60968-x. [DOI] [PubMed] [Google Scholar]
- 2.Simpson R T. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 3.Allan J, Hartman P G, Crane-Robinson C, Aviles F X. Nature (London) 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 4.Staynov D Z, Crane-Robinson C. EMBO J. 1988;7:3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pehrson J R. Proc Natl Acad Sci USA. 1989;86:9149–9153. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 7.Goytisolo F A, Gerchman S-E, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas J O. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes J J, Wolffe A P. Proc Natl Acad Sci USA. 1993;90:6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes J J, Pruss D, Wolffe A P. Proc Natl Acad Sci USA. 1994;91:7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ura K, Hayes J J, Wolffe A P. EMBO J. 1995;15:3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis E N, Wolffe A P. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 12.Hayes J J. Biochemistry. 1996;35:11931–11937. doi: 10.1021/bi961590+. [DOI] [PubMed] [Google Scholar]
- 13.Travers A A, Muyldermans S V. J Mol Biol. 1996;257:486–491. doi: 10.1006/jmbi.1996.0178. [DOI] [PubMed] [Google Scholar]
- 14.Muyldermans S, Travers A A. J Mol Biol. 1994;235:855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- 15.Crane-Robinson C. Trends Biochem Sci. 1997;22:75–77. doi: 10.1016/s0968-0004(97)01013-x. [DOI] [PubMed] [Google Scholar]
- 16.Arents G, Moudrianakis E N. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arents G, Burlingame R W, Wang B-C, Love W E, Moudrianakis E N. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 19.Simpson R T, Stafford D W. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong F, Hansen J C, van Holde K E. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pennings S, Meersseman G, Bradbury E M. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 22.Meersseman G, Pennings S, Bradbury E M. J Mol Biol. 1991;220:89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- 23.Adams C C, Workman J L. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doenecke D, Tönjes R. J Mol Biol. 1986;187:461–464. doi: 10.1016/0022-2836(86)90446-8. [DOI] [PubMed] [Google Scholar]
- 25.Simpson R T, Thoma F, Brubaker J M. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 26.Tatchell K, van Holde K E. Biochemistry. 1977;16:5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- 27.Simon R H, Felsenfeld G. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Bavykin S G, Usachenko S I, Zalensky A O, Mirzabekov A D. J Mol Biol. 1990;212:495–511. doi: 10.1016/0022-2836(90)90328-J. [DOI] [PubMed] [Google Scholar]
- 31.Wong J, Li Q, Levi B-Z, Shi Y-B, Wolffe A P. EMBO J. 1997;23:7130–7145. doi: 10.1093/emboj/16.23.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. J Mol Biol. 1995;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]