Abstract
Although nucleic acid polymerases from different families show striking similarities in structure, they maintain stringent specificity for the sugar structure of the incoming nucleoside triphosphate. The Klenow fragment of E. coli DNA polymerase I selects its natural substrates, deoxynucleotides, over ribonucleotides by several thousand fold. Analysis of mutant Klenow fragment derivatives indicates that discrimination is provided by the Glu-710 side chain which sterically blocks the 2′-OH of an incoming rNTP. A nearby aromatic side chain, at position 762, plays an important role in constraining the nucleotide so that the Glu-710 “steric gate” can be fully effective. Even with the E710A mutation, which is extremely permissive for addition of a single ribonucleotide to a DNA primer, Klenow fragment does not efficiently synthesize pure RNA, indicating that additional barriers prevent the incorporation of successive ribonucleotides.
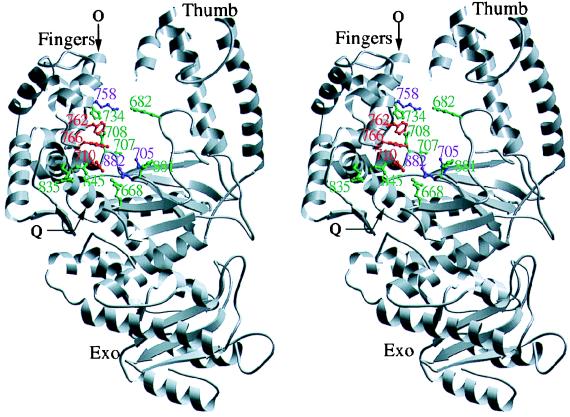
It is evident from structural studies that nucleic acid polymerases constitute a superfamily sharing a common three-dimensional architecture of the polymerase domain (for reviews see refs. 1–3). This view tends to deemphasize divisions based on whether a polymerase is DNA- or RNA-directed and whether it makes a DNA or an RNA product, distinctions that, though important, probably do not reflect fundamental mechanistic differences. The characteristic polymerase domain fold has been compared with a half-open right hand, with subdomains described as palm, fingers, and thumb (Fig. 1). The palm subdomain, which forms the floor of the polymerase cleft, contains many of the important active site residues, including a cluster of highly conserved carboxylates that coordinate two divalent metal ions in the appropriate positions to catalyze the phosphoryl transfer reaction (1, 6, 7). The incoming nucleotide is nestled against the fingers side of the cleft; movement of the fingers subdomain probably accompanies formation of the ternary complex, generating the appropriate arrangement of interacting side chains by an induced fit mechanism (7).
Figure 1.
A stereo view of the Klenow fragment of DNA polymerase I (Kf pol) structure (4). The separate 3′-5′ exonuclease domain is indicated, as are the fingers and thumb subdomains that form the sides of the polymerase cleft. Within the polymerase domain are shown two long helices: O, on the fingers subdomain, and Q, connecting fingers and palm subdomains. The side chains shown in green indicate the positions of mutations that had little or no effect on dNTP/rNTP discrimination (Table 1); those shown in red correspond to mutations whose phenotypes are discussed in the text. Side chains shown in purple are the important catalytic residues, Asp-705, Asp-882, and Lys-758, which could not be screened because the relevant mutant proteins had too little activity. This figure was made by Jimin Wang, using molscript (5).
Our earlier studies of mutant derivatives of DNA polymerase I Klenow fragment (Kf pol) showed that the O-helix in the fingers subdomain (Fig. 1) is particularly important in binding an incoming dNTP (8), and the ternary complex structure of T7 DNA polymerase has confirmed many of our predictions (7). The sugar portion of the nucleotide is bound close to the C-terminal region of the O-helix, and the identification of mutations affecting sugar specificity has been used to explore interactions that are important in recognizing a deoxyribose sugar. Studies on the incorporation of dideoxynucleotides by polymerases of the Pol I family have implicated Phe-762 and Glu-710 in monitoring the 3′ substituent (9–11). In this work we have used ribonucleotide incorporation as a criterion to identify side chains that monitor the 2′ position.
A similar study identified an aromatic side chain in the active site of reverse transcriptases as providing a “steric gate” that prevents incorporation of ribonucleotides (12), and the opposite phenotype, allowing an RNA polymerase to use deoxynucleotides, has also been described (13). In both cases the relevant side chains are in the same area of the polymerase cleft as described above for Pol I family polymerases, implicating this part of the structure as containing important determinants that differentiate DNA from RNA polymerases.
MATERIALS AND METHODS
Substrates.
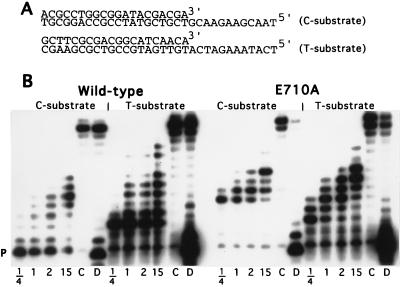
Ultra-pure dNTPs and rNTPs were from Pharmacia LKB Biotech. Cordycepin (3′-dATP), and its C and U analogs, originally from Pharmacia, were kindly provided by Stanley Tabor of Harvard University (3′-dGTP was not available). Radiolabeled nucleotides were purchased from Amersham Life Science. The synthetic DNA duplex substrates for the pre-steady-state kinetic measurements (see Fig. 4A) have been described previously (11).
Figure 4.
Addition of successive rNTPs. (A) The two DNA substrates used in this experiment and in the kinetic studies. (B) Extension of the 20-mer primers (P) by wild-type and E710A Kf pol in the presence of all four rNTPs at 83 μM. Samples were taken at the indicated times (hr); the 15-hr sample was supplemented with fresh rNTPs (100 μM) and polymerase after 2 hr of incubation. C denotes a portion of the 2-hr sample that was chased with dNTPs (60 μM) and wild-type Kf pol (≈3 μM) for 13 hr; an aliquot of this reaction mixture was then digested with piperidine (D).
Mutant Proteins.
Mutant derivatives of Kf pol were constructed and purified as described elsewhere (14, 15). Biochemical properties of the majority of these proteins have been reported previously (8, 11, 14, 16, 17). All Kf pol derivatives used in this study carry the D424A mutation, which eliminates the 3′-5′ exonuclease activity (18).
Screening for Discrimination Between dNTPs and rNTPs or 3′-dNTPs.
The assay for dNTP/rNTP discrimination was based on the DNA sequencing procedure developed by Barnes (19), except that Mg2+ (at 2 mM) was the metal cofactor for the polymerase reaction. The DNA substrate consisted of the 20-mer oligonucleotide that forms the primer strand of the C-substrate (Fig. 4A), 32P-labeled at the 5′ terminus and annealed to a complementary M13 template. Two microliters of a solution containing ≈15 nM DNA (as primer termini) in 10 mM Tris⋅HCl, pH 7.5/6 mM MgCl2 was mixed with 2 μl of a mix containing all four dNTPs and a single rNTP (giving final nucleotide concentrations as indicated in the legend of Fig. 2). Primer extension was initiated by adding 2 μl of a Kf pol derivative, diluted to about 2 μM in 10 mM Tris⋅HCl, pH 7.5/10 mM DTT, and the reaction mixture was incubated at 37°C for 90 min. A sample (2 μl) was taken to ascertain that the primer had been elongated sufficiently during the initial incubation. To the remaining reaction mix (about 4 μl) was added 2 μl of a solution containing 250 μM of each of the four dNTPs and 2–3 μM wild-type Kf pol. The mixture was incubated for 10 min at 37°C and a 2 μl sample was taken to check that all products had been chased into high molecular weight DNA, so that bands seen after the subsequent alkali digestion could be attributed unambiguously to ribonucleotide incorporation. The remainder of the reaction mixture was lyophilized, redissolved in 1 M piperidine (10 μl), and incubated at 67°C for 3–4 hr so as to cleave at the ribonucleotide positions. After the cleavage reaction, the piperidine was removed by repeated lyophilization from water (20 μl) and the product was dissolved in 4 μl of gel-loading dyes [67% (vol/vol) deionized formamide, 10 mM EDTA, and 0.007% each of bromophenol blue and xylene cyanol FF]. Samples were fractionated on a denaturing 6% polyacrylamide gel.
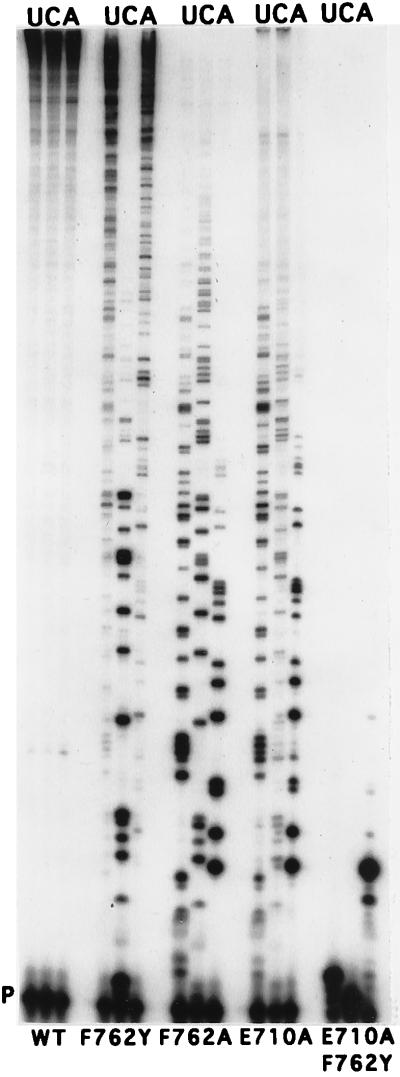
Figure 2.
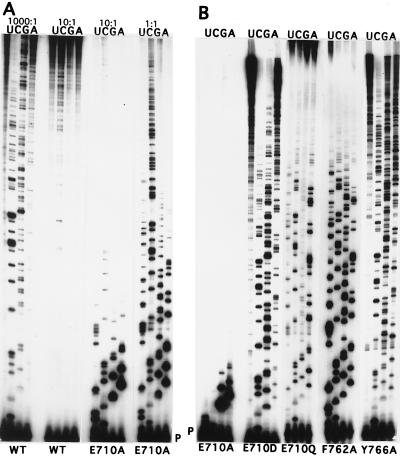
Gel assay of dNTP/rNTP discrimination. (A) Comparison of wild-type and E710A Kf pol at various rNTP:dNTP ratios, indicated above each set of four lanes. A ratio of 1000:1 corresponded to 1.5 mM rNTP (U, C, G, or A, as indicated) and 1.5 μM of each dNTP in the reaction. For 10:1 and 1:1 ratios, dNTPs were present at 13 μM. P indicates the position of the labeled primer. Comparison of the products shown here with samples taken before alkali digestion confirmed that the bands seen represent bona fide ribonucleotide incorporation. (B) Comparison of various mutant derivatives of Kf pol under the same conditions as in A, except that the rNTP:dNTP ratio was 200:1, with each of the four dNTPs at 1.5 μM. The data in B were taken from several gels; hence, corresponding bands are not necessarily in identical positions.
The assay for discrimination between dNTPs and 3′-dNTPs was based on the Sanger sequencing method (20), as described previously for 2′,3′-dideoxy (dd)NTP/dNTP competition (11), with nucleotide mixes containing 2 μM of each of the four dNTPs and 40 μM of a single 3′-dNTP.
Incorporation of Successive Ribonucleotides.
The two DNA substrates illustrated in Fig. 4A were used. Five microliters of a solution containing ≈15 nM DNA (as primer termini) in 10 mM Tris⋅HCl, pH 7.5/6 mM MgCl2 was mixed with 5 μl of a solution containing all four rNTPs at 250 μM. Primer extension was initiated by adding 5 μl of wild-type or E710A Kf pol, diluted to 1.5 μM (wild-type) or 6 μM (E710A) in 10 mM Tris⋅HCl, pH 7.5/10 mM DTT, and the reaction mixture was incubated at 22°C. The samples taken (see Fig. 4 legend) were fractionated on a denaturing 8% polyacrylamide gel.
Kinetic Measurements.
Single-turnover measurements for rCTP and rUTP substrates were carried out as described previously for dNTPs and ddNTPs (11).
RESULTS
Discrimination Between dNTPs and rNTPs by Kf pol Mutants.
To identify those residues in Kf pol that may be close to the 2′ position of the incoming dNTP we compared the ability of wild-type and mutant derivatives to distinguish between dNTPs and rNTPs. We focused on residues within the polymerase active site region (Fig. 1), especially those close to Phe-762 and Glu-710, previously shown to be important in selecting for the hydroxyl at the 3′ position of the nucleotide (11). Throughout this study, Mg2+ was used as the metal ion cofactor. To avoid complications caused by the 3′-5′ exonuclease activity, all Kf pol derivatives used in this study carried the exonuclease-deficient mutation D424A (18). For simplicity, we describe each protein in terms of the genotype of the polymerase domain; thus, the D424A control is referred to as wild-type, and E710A denotes the double mutation D424A, E710A.
The ability of wild-type and mutant derivatives of Kf pol to discriminate between rNTPs and dNTPs was first assessed qualitatively. Primer extension was carried out with dNTP and rNTP competing as substrates for the polymerase; ribonucleotide positions were then identified by alkali cleavage (19, 21). A reduced level of discrimination between dNTP and rNTP by a mutant derivative will result in a higher probability of rNMP incorporation and a greater yield of short oligonucleotide products after alkali cleavage, compared with the reaction catalyzed by the wild-type protein. At a ratio of rNTP to dNTP of 1000:1, wild-type Kf pol used rCTP and rGTP to a significant extent, but use of rUTP and rATP was barely detectable (Fig. 2A), in good agreement with Ide et al. (21). The most dramatic decrease in dNTP/rNTP discrimination was seen with the E710A mutant derivative of Kf pol, which gave a substantial level of rNMP incorporation at a 1:1 ratio of rNTP to dNTP (Fig. 2A), and even at ratios as low as 0.1:1 (data not shown). For comparison, wild-type Kf pol gave hardly any rNMP insertion at a 10:1 ratio of rNTP to dNTP (Fig. 2A). Other substitutions at Glu-710 (E710D and E710Q) used rNTPs more than wild type but less than the E710A mutant derivative (Fig. 2B). The E710D polymerase showed the wild-type order of preference for rNTPs (C > G ≫ A, U), whereas the E710A and E710Q derivatives accepted all four rNTPs at roughly similar frequencies.
Ribonucleotide incorporation was also affected by some substitutions in the region around the C terminus of the O-helix in the fingers subdomain (Fig. 1). The F762A mutant protein showed a level of rNMP insertion intermediate between the levels of the E710D (or Q) and the E710A mutant derivatives (Fig. 2B); moreover, all four rNTPs were used to similar extents. The conservative F762Y substitution at this same position gave a polymerase indistinguishable from wild-type Kf pol in the dNTP/rNTP competition assay (data not shown). A variety of phenotypes were observed for mutations at Tyr-766. The Y766A derivative gave a level of rCMP incorporation very similar to that of wild type, but, unlike wild-type Kf pol, it also incorporated rGMP and rAMP (but not rUMP) to an extent similar to that of rCMP (Fig. 2B). The dNTP/rNTP discrimination by theY766S mutant protein was similar to that of wild-type Kf pol, whereas the Y766F mutation resulted in slightly enhanced discrimination against ribonucleotides (data not shown). In this same region of the protein, the R835L mutation gave essentially the same result as Y766A, and N845A was similar, except that use of rUTP was increased so that the incorporation of all four rNMPs was approximately equal (data not shown). The other mutant proteins that were tested (R668A, R682A, S707A, Q708A, H734A, and H881A, see Fig. 1) behaved essentially the same as wild-type Kf pol; given the qualitative nature of the assay, however, we cannot exclude the possibility that some of these mutations might have small effects on rNTP use.
Kinetic Analysis of Ribonucleotide Incorporation.
To investigate the underlying causes of the changes in dNTP/rNTP discrimination associated with mutations at Glu-710 and Phe-762, we studied the kinetics of the polymerase reaction with rNTP substrates. Two different synthetic DNA duplexes were used so as to study the incorporation of C and T(U) nucleotide derivatives. The polymerase reaction was limited to a single turnover by having enzyme in excess over the DNA substrate so that all the DNA was enzyme-bound at the start of the experiment. The observed rate then reflects the rates of steps up to and including phosphoryl transfer and is limited either by the phosphoryl transfer step or by the preceding noncovalent “conformational change” (22). By plotting rate as a function of nucleotide concentration, the dissociation constant (Kd) for binding of the nucleotide to form the ternary complex, and the maximal reaction rate (kcat) were determined (Table 1).
Table 1.
Pre-steady-state kinetic constants for use of dNTPs and rNTPs by Kf pol and mutant derivatives
| Protein* | Kd, μM† | kcat, s−1† | kcat/Kd | Kd, μM† | kcat, s−1† | kcat/Kd | Selection‡ |
|---|---|---|---|---|---|---|---|
| dCTP | rCTP | dC/rC | |||||
| Wild type | 9.6 ± 2.3 | 75 ± 13 | 7.8 | 21 ± 7 | (4.7 ± 2.5) × 10−2 | 2.3 × 10−3 | 3,400 |
| E710A | 18 ± 7 | 4.2 ± 0.4 | 0.23 | 3.5 ± 1.1 | 0.2 ± 0.1 | 5.4 × 10−2 | 4.3 |
| E710D | 7.7 ± 2.7 | (1.8 ± 0.1) × 10−2 | 2.4 × 10−3 | ||||
| E710Q | 11 ± 2 | (2.1 ± 0.2) × 10−2 | 2.0 × 10−3 | ||||
| F762A | 110 ± 20 | 19 ± 5 | 0.17 | 140 ± 50 | (8.9 ± 3.4) × 10−2 | 6.6 × 10−4 | 260 |
| dTTP | UTP | dT/rU | |||||
| Wild type | 20 ± 6 | 87 ± 6 | 4.3 | 220 ± 50 | (5.6 ± 1.0) × 10−4 | 2.5 × 10−6 | 1.7 × 106 |
| E710A | 35 ± 7 | 3.4 ± 0.3 | 0.1 | 85 ± 3 | 0.26 ± 0.02 | 3.0 × 10−3 | 33 |
All the proteins in this study carry the D424A mutation and are therefore deficient in 3′-5′ exonuclease activity.
Kinetic parameters were from at least three independent determinations and are reported as mean ± SD. The dNTP data are from our previous work (11).
The selection factor for dNTP over rNTP is given by the ratio of the efficiencies (kcat/Kd) for dNTP and rNTP addition.
Wild-type Kf pol discriminates against rCTP by 3,000-fold and against rUTP by more than a million-fold (Table 1). In both cases, the discrimination is almost entirely attributable to a very low kcat for rNMP incorporation. Relative to wild-type Kf pol, the E710A mutation reduces the discrimination against the rNTP substrate by almost 1,000-fold for rCTP and by 105-fold for rUTP; this is the combination of a decrease in the efficiency of using the natural (dNTP) substrate and an improvement in the efficiency of ribonucleotide insertion (Table 1). As we have noted previously (11), the slower reaction with dNTPs is at least partly attributable to weaker Mg2+ binding by the E710A protein, so that the standard conditions (2 mM Mg2+) used for the kinetic measurements are not at the optimal Mg2+ concentration. The E710D and E710Q mutant derivatives had kinetic constants similar to those of wild-type Kf pol when using rNTPs (Table 1). Given that the steady-state kinetic parameters for E710D and E710Q (Table 2) (11) indicate decreased efficiency in handling the natural dNTP substrate, the data are broadly consistent with the moderate loss of discrimination against rNTPs observed in the gel assay. The kinetic data for the F762A protein, giving a dCTP/rCTP selection factor intermediate between the factors for wild-type and E710A, also agree well with the more qualitative assessment from the gel assay. The F762A mutation causes a substantial defect in binding both dCTP and rCTP; the decreased discrimination against rCTP occurs primarily because the rate of rCMP addition by F762A is slightly faster than with wild type, even though use of the natural (dCTP) substrate is slower.
Table 2.
Effect of mutations at Glu-710 of Kf pol on steady-state kinetic constants for dTTP addition
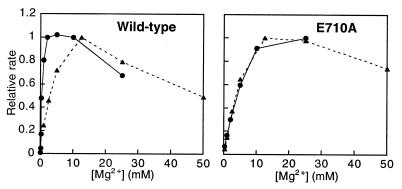
To investigate whether divalent metal ions may be involved in discrimination between dNTPs and rNTPs, we measured the rate of rCMP incorporation as a function of Mg2+ concentration. For both wild-type Kf pol and the E710A mutant protein, the Mg2+ optimum was around 12.5 mM, and Mg2+ concentrations above about 25 mM were inhibitory (Fig. 3). This contrasts with deoxynucleotide incorporation (exemplified by dTTP), where the E710A mutant protein has a much higher Mg2+ optimum than does wild type (11).
Figure 3.
Dependence of reaction rate on Mg2+ concentration for wild-type and E710A Kf pol. The single-turnover rate for rCMP addition to the C-substrate (Fig. 4A) was measured at a series of Mg2+ concentrations from 0.25 to 50 mM, with rCTP at 0.2 μM (▴, broken lines). (The Mg2+ optimum was essentially the same when the experiment was repeated at a different rCTP concentration.) The Mg2+ dependence for dTMP incorporation by these two proteins (•, solid lines) is taken from steady-state measurements; as we have argued previously (11), the comparison of pre-steady-state and steady-state data appears to be justified in this case. For each plot, the maximal rate is arbitrarily set at 1.
Addition of Successive rNTPs.
Because the E710A mutation relaxes discrimination against rNTPs at the nucleotide insertion step, we investigated whether it would allow Kf pol to function as a true RNA polymerase, adding successive rNMPs to a polynucleotide chain. The abilities of wild-type and E710A Kf pol to extend DNA primers with rNTPs were compared, using the synthetic oligonucleotide T- and C-substrates (Fig. 4A), each of which has a 12-nucleotide 5′ extension to serve as a template. Although the E710A polymerase adds a single rNMP more rapidly than wild type, the overall profiles for the two enzymes were very similar, showing that both were extremely inefficient at synthesizing a product consisting entirely of ribonucleotides (Fig. 4B). As with any primed synthesis reaction on a heteropolymeric sequence, the product bands were not of uniform intensity, reflecting the influence of local sequence context on polymerase pausing. A general trend with both substrates was that synthesis slowed as more rNMPs were added, leading to substantial accumulation of primers having 4 to 7 residues of RNA; nevertheless, these molecules could be chased into full-length products with dNTPs (C lanes). Evidence that the products represented bona fide RNA synthesis (rather than use of dNTPs present as minor contaminants in the reaction mix) was provided by the differences in electrophoretic mobility when the RNA synthesis lanes were compared with the dNTP-chased samples, and by the demonstration that almost all of the chased product could be degraded with alkali to regenerate the starting material (D lanes). (The small amount that was not degraded probably corresponds to unextended primer remaining after 2 hr, which, when chased, would give rise to a product consisting entirely of deoxynucleotides.)
3′-dNTP Analogs.
We measured the use by Kf pol of 3′-dNTPs, chain terminators that combine the noncanonical features of rNTPs and ddNTPs. The ability of selected mutant Kf pol derivatives to accept 3′-dNTPs as substrates (Fig. 5) agreed with the predictions from our studies of rNTP and ddNTP substrates (11). The F762A and E710A mutations reduced the discrimination against 3′-dNTPs, consistent with their tolerance of both rNTPs and ddNTPs. The F762Y protein, which uses ddNTPs efficiently but discriminates strongly against rNTPs, gave significant incorporation of the 3′-deoxy analog, but less than F762A and E710A. Combining F762Y, the most permissive mutation for ddNTPs, with E710A, the most permissive for rNTPs, gave a doubly mutant protein that discriminated very poorly against 3′-dNTPs.
Figure 5.
Gel assay of dNTP/3′-dNTP discrimination by wild-type and various mutant derivatives of Kf pol. A single 3′-dNTP (U, C, or A, as indicated), which acts as a chain terminator, competes with the corresponding dNTP during elongation of the primer (P). The ratio of 3′-dNTP to dNTP was 20:1.
DISCUSSION
A Steric Model for dNTP/rNTP Discrimination.
Polymerases show exquisite specificity for their nucleotide substrate, not only selecting the correct base in response to template coding information but also choosing the correct sugar structure. For DNA polymerases the problem is particularly acute because rNTPs, a possible competing substrate, are present in vivo at about 10 times the concentration of dNTPs (23). Here we have shown that Kf pol discriminates against rNTPs by a factor of at least 3,000-fold (the value for rCTP, the best substrate of the four rNTPs). The most obvious difference between ribo- and deoxyribonucleotides is the presence of the 2′-hydroxyl in the ribonucleotide, so that selectivity could be achieved by steric exclusion of the 2′-OH. Discrimination against rNTPs is manifested almost entirely in kcat; therefore unfavorable interactions with the 2′-OH must be felt more in the transition state than in ground state nucleotide binding.
Our data for mutant derivatives of Kf pol are consistent with a steric exclusion model, and they suggest that Glu-710 obstructs the insertion of rNTPs. Substitution of the much smaller Ala side chain gives a polymerase that discriminates very poorly against rNTPs, whereas intermediate-sized substitutions cause only a moderate decrease in dNTP/rNTP discrimination. The properties of the F762A mutant protein also support the steric model. The weaker nucleotide binding and lower selectivity of the F762A protein against dideoxynucleotides (Table 1) (8, 11) have led us to conclude that the Phe-762 side chain plays a major role in positioning the sugar moiety of the incoming nucleotide. In the absence of the aromatic side chain, an incoming rNTP is less rigidly constrained (reflected in the high Kd value), and consequently the 2′-OH is better able to avoid a steric clash with Glu-710. Defects in binding and positioning of the substrate by the F762A polymerase presumably account for dCMP addition being 1/4 as fast as with wild-type. By contrast, the rate of rCMP addition by F762A is 2-fold faster than wild type. Consistent with the steric interpretation, the F762Y mutation, preserving the aromatic side chain, does not alleviate discrimination against rNTPs.
The ability of certain Kf pol mutants to use 3′-dNTPs, which combine the noncanonical aspects of dideoxy- and ribonucleotides, is fully consistent with the interactions that we have proposed as responsible for ensuring the correct structure at the 2′ and 3′ positions of the sugar. The present study postulates Glu-710 as the “steric gate” that obstructs an incoming rNTP, and our previous work implicated Phe-762 and Glu-710 in setting up an interaction that selects for the dNTP 3′-OH (11). Consistent with expectations, the use of 3′-dNTPs was enhanced by the E710A mutation, which is involved in both 2′ and 3′ recognition, and by the F762A mutation which, by binding the sugar moiety less rigidly, allows the nucleotide to evade the interactions that monitor the 2′ and 3′ substituents. The F762Y mutation, which enhances ddNTP use by providing an interaction that compensates for the lack of a 3′-OH, was less permissive than F762A unless combined with E710A to remove the steric obstruction at the 2′ position.
How Does Glu-710 Function as a Steric Gate?
The structure of T7 DNA polymerase in a complex with its two substrates suggests a way in which Glu-710 could provide selectivity against ribonucleotides, but it leaves some questions unresolved. In T7 DNA polymerase the glutamate equivalent to Glu-710 of Kf pol is hydrogen-bonded to the γ-OH of the invariant tyrosine at the C terminus of the O-helix (equivalent to Tyr-766), with the consequence that the aliphatic carbons of the Glu side chain are positioned such that they could exclude the 2′-OH of an incoming rNTP (7). This interaction readily accounts for the phenotypes of E710A and E710D. Ala is much smaller than Glu and cannot make a hydrogen bond with Tyr-766, leaving plenty of space for the 2′-OH. Asp is also smaller and, if it made a hydrogen bond, would likely be shifted into a slightly different position from Glu-710, relieving somewhat the steric constraint. However, while the observed interaction between the homologs of Glu-710 and Tyr-766 is satisfying in that it suggests functions for two side chains that are invariant among all DNA polymerases of the Pol I family (11, 24), it does not convincingly account for the phenotypes of some of the other Kf pol mutations. First, it is hard to understand why E710Q should affect discrimination against rNTPs; Gln is isosteric with Glu and could hydrogen-bond with Tyr-766; thus it too should block the 2′-OH. Second, the mutation Y766F, which would disrupt the proposed hydrogen bond with Glu-710, not only has no effect on rNMP incorporation but also fails to affect the kinetic parameters for dNTP substrates (11, 17). These observations suggest that the 710–766 hydrogen bond observed in the crystal structure may be either functionally unimportant or a result of using a dideoxynucleotide in the ternary complex.
The behavior of mutant proteins with substitutions of Glu-710 suggests to us that the important attribute of Glu-710 is its ability to serve as a metal ligand, since E710D is a more effective polymerase than E710Q (Table 2). This interpretation fits with our earlier suggestion that a metal ion associated with Glu-710 provides selectivity for the 3′-OH of the incoming nucleotide (11). The Mg2+ dependence of ribonucleotide incorporation also supports the idea that a metal ion could be bound somewhere in the vicinity of Glu-710 and the sugar 2′ and 3′ positions. Wild-type Kf pol has a higher Mg2+ optimum when using rNTPs than when using dNTPs, implying that the sugar 2′-OH interferes with metal binding. Compared with wild-type, the E710A mutant protein has a higher Mg2+ optimum with dNTP substrates but the same Mg2+ optimum with rNTP substrates, suggesting that Glu-710 is a metal ligand when the substrate is a dNTP but is made irrelevant by the disruption caused by a 2′-OH substituent. Although we cannot rule out an indirect effect of substitutions at 710 on metal binding, the scenario we propose raises an interesting possibility. If a metal ion is indeed bound to Glu-710, then steric exclusion of the 2′-OH might involve a clash with the metal ion, rather than the Glu-710 side chain. Thus a single metal ion could mediate discrimination at both the 2′ and 3′ positions of the dNTP, selecting the 2′-H by steric exclusion and the 3′-OH by making an interaction.
Base Specificity in dNTP/rNTP Discrimination.
A curious feature of our data is that some mutant Kf pol derivatives, such as E710A and F762A, used all four rNTPs to a roughly similar extent whereas others, such as E710D, followed the wild-type pattern of using rCTP and rGTP more readily than rATP and rUTP (Fig. 2, Table 1) (21). Although strong discrimination between rUTP and dTTP could be attributed to the absence of the 5-methyl substituent on uracil in addition to the difference in sugar structure, this does not account for the similarly strong discrimination against rATP. We therefore believe that the base specificity reflects some characteristic of A⋅T(U) versus C⋅G base pairs. Donlin and Johnson (25) have pointed out that polymerases might be expected to have a mechanism for selectively stabilizing an incoming A or T nucleotide (relative to C or G), because this would explain the approximately equal dissociation constants for all four dNTPs regardless of the number of hydrogen bonds formed with the template base. If such a mechanism exists, our data suggest that it is compromised by the use of ribonucleotide substrates with the wild-type enzyme, but can be restored, to various extents, by certain mutations.
Ribonucleotide Discrimination by Other Polymerases.
Examination of protein structures and sequences indicates that other DNA polymerases may have conserved residues positioned such that they can discriminate by steric exclusion against a 2′ substituent on the incoming nucleotide. These ideas have been tested experimentally in the case of Moloney murine leukemia virus reverse transcriptase (MMLV-RT). Model building, based on the structure of a fragment of the polymerase domain of MMLV-RT, suggested that the side chain of Phe-155 would clash with the 2′-OH of a ribonucleotide (26); mutation to a smaller side chain (valine) allowed the use of rNTPs (12). Superficially, the F155V mutation in reverse transcriptase appears analogous to the E710A mutation in Kf pol, but the kinetic properties of the two proteins differ significantly. Discrimination by MMLV-RT against rNTPs is manifested about equally in Km and kcat for nucleotide incorporation. The F155V mutation alleviates the Km discrimination (presumably corresponding to ground-state binding), but the rate of reaction with rNTP substrates is only marginally improved relative to wild type and remains 1/100 as fast as the reaction with dNTPs, suggesting that there are additional ways in which MMLV-RT monitors the sugar structure of the incoming nucleotide. Wild-type Kf pol, on the other hand, discriminates very little against rNTPs in binding (Table 1); as discussed above, the steric consequences of the 2′-OH substituent are manifested in one of the transition states.
The two “discriminator” side chains identified thus far (Glu-710 in Kf pol and Phe-155 in MMLV-RT) occupy identical positions on the primary sequence, being five residues C-terminal to the invariant active-site aspartate of motif A (27, 28). Both amino acids are highly conserved in their respective polymerase families, the glutamate being invariant in the Pol I family (11) and the Phe-155 position being occupied by Phe or Tyr in reverse transcriptases (29). In the Pol α family the corresponding sequence position is an invariant tyrosine (24), and model-building, using the structure of bacteriophage RB69 DNA polymerase, suggests that this side chain could serve a similar discriminator role (30). Because the motif A region of the polymerase palm subdomain is essentially superimposable in the structures of DNA polymerases from these three families (30), the close correspondence between sequence position, location in three-dimensional space, and probable function is perhaps not too surprising. The DNA polymerase β structure, though not superimposable on the other DNA polymerases, also has an aromatic side chain (Tyr-271) that makes close contact with the sugar 2′ position (6). Consistent with the expectations of the steric model, RNA polymerases have smaller side chains at the corresponding sequence position (28). Thus, similar steric mechanisms appear likely to operate in a variety of DNA polymerases, although the Pol I family may be unusual in using a carboxylate instead of an aromatic side chain.
Barriers to Incorporation of Successive Ribonucleotides.
Although mutations in the discriminator residues identified in Kf pol and MMLV-RT remove barriers to the addition of a single ribonucleotide, they do not cause a substantial improvement in the ability to synthesize a product composed entirely of the noncanonical substrate. A similar observation was reported with an altered-specificity mutation that allows T7 RNA polymerase to use dNTPs (13, 31). The additional barriers that must be overcome when a DNA polymerase adds successive ribonucleotides could be of two general types: difficulty in accommodating a 2′-OH substituent on the primer-terminal residue at the active site, and problems with binding an RNA⋅DNA duplex in the primer–template (product) site. The preferred conformation for an RNA⋅DNA duplex is an A-like helix (32, 33), whereas a DNA⋅DNA duplex, the normal product of a DNA polymerase, though favoring the B form, is able to assume either A- or B-form conformations. When DNA is bound to DNA polymerases the duplex tends to adopt a more A-like configuration close to the active site (6, 7, 34–36), suggesting that a small stretch of ribonucleotides at the primer terminus might be tolerated reasonably well, but problems could arise if the newly synthesized RNA reached that part of the product binding site that is better suited for binding a B-form duplex. Synthesis of a purely RNA product by Kf pol becomes increasingly slow after about four or five rNTPs have been added, consistent with the above scenario. Identifying the additional barriers that preclude efficient addition to a ribonucleotide primer terminus and/or synthesis of an exclusively ribonucleotide product could shed light on the functional and evolutionary relationships between DNA and RNA polymerases.
Acknowledgments
We thank Tom Steitz, Joe Jaeger, Soo Hyun Eom, and Jimin Wang for their insights into polymerase structures, Tom Ellenberger for helpful discussions and for communicating results prior to publication, Jimin Wang for help with figures, and Xiaojun Chen Sun for expert technical assistance. This work was supported by National Institutes of Health Grant GM-28550, including a Minority Supplement to M.A.
ABBREVIATIONS
- Kf pol
Klenow fragment of DNA polymerase I
- ddNTP
2′,3′-dideoxynucleoside triphosphate
- MMLV-RT
Moloney murine leukemia virus reverse transcriptase
References
- 1.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 2.Joyce C M, Steitz T A. J Bacteriol. 1995;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa R. Trends Biochem Sci. 1996;21:186–190. [PubMed] [Google Scholar]
- 4.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 5.Priestle J P. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 6.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 7.Doublié S, Tabor S, Long A, Richardson C C, Ellenberger T. Nature (London) 1997;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 8.Astatke M, Grindley N D F, Joyce C M. J Biol Chem. 1995;270:1945–1954. doi: 10.1074/jbc.270.4.1945. [DOI] [PubMed] [Google Scholar]
- 9.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizrahi V, Huberts P. Nucleic Acids Res. 1996;24:4845–4852. doi: 10.1093/nar/24.24.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Astatke, M., Grindley, N. D. F. & Joyce, C. M. (1998) J. Mol. Biol. 278, in press. [DOI] [PubMed]
- 12.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa R, Padilla R. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polesky A H, Steitz T A, Grindley N D F, Joyce C M. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 15.Joyce C M, Derbyshire V. Methods Enzymol. 1994;262:3–13. doi: 10.1016/0076-6879(95)62003-6. [DOI] [PubMed] [Google Scholar]
- 16.Polesky A H, Dahlberg M E, Benkovic S J, Grindley N D F, Joyce C M. J Biol Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 17.Carroll S S, Cowart M, Benkovic S J. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 18.Derbyshire V, Freemont P S, Sanderson M R, Beese L, Friedman J M, Joyce C M, Steitz T A. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 19.Barnes W M. J Mol Biol. 1978;119:83–99. doi: 10.1016/0022-2836(78)90271-1. [DOI] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ide H, Yagi R, Yamaoka T, Kimura Y. Nucleic Acids Symp Ser. 1993;29:133–134. [PubMed] [Google Scholar]
- 22.Dahlberg M E, Benkovic S J. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg A, Baker T A. DNA Replication. San Francisco: Freeman; 1992. [Google Scholar]
- 24.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donlin M J, Johnson K A. Biochemistry. 1994;33:14908–14917. doi: 10.1021/bi00253a030. [DOI] [PubMed] [Google Scholar]
- 26.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 27.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 28.Joyce C M. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Sattar A K M A, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Beaudry A, McSwiggen J, Sousa R. Biochemistry. 1997;36:13718–13728. doi: 10.1021/bi971609o. [DOI] [PubMed] [Google Scholar]
- 32.Fedoroff O Y, Salazar M, Reid B R. J Mol Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 33.Hall K B, McLaughlin L W. Biochemistry. 1991;30:10606–10613. doi: 10.1021/bi00108a002. [DOI] [PubMed] [Google Scholar]
- 34.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eom S H, Wang J, Steitz T A. Nature (London) 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 36.Kiefer J R, Mao C, Braman J C, Beese L S. Nature (London) 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]