Abstract
Two related oncogenes, TCL-1 and MTCP-1, are overexpressed in T cell prolymphocytic leukemias as a result of chromosomal rearrangements that involve the translocation of one T cell receptor gene to either chromosome 14q32 or Xq28. The crystal structure of human recombinant MTCP-1 protein has been determined at 2.0 Å resolution by using multiwavelength anomalous dispersion data from selenomethionine-enriched protein and refined to an R factor of 0.21. MTCP-1 folds into a compact eight-stranded β barrel structure with a short helix between the fourth and fifth strands. The topology is unique. The structure of TCL-1 has been predicted by molecular modeling based on 40% amino acid sequence identity with MTCP-1. The identical residues are clustered inside the barrel and on the surface at one side of the barrel. The overall structure of MTCP-1 superficially resembles the structures of proteins in the lipocalin family and calycin superfamily. These proteins have diverse functions, including transport of retinol, fatty acids, chromophores, pheromones, synthesis of prostaglandin, immune modulation, and cell regulation. However, MTCP-1 differs in the topology of the β strands. The structural similarity suggests that MTCP-1 and TCL-1 form a unique family of β barrel proteins that is predicted to bind small hydrophobic ligands and function in cell regulation.
Chromosomal rearrangements in the T cell malignancies observed in T cell prolymphocytic leukemias, and in patients with Ataxia telangiectasia involve the translocation of one T cell receptor gene to either chromosome 14q32 or Xq28 (1–3). These genetic aberrations juxtapose cellular protooncogenes to enhancer elements leading to deregulation of oncogene expression (4). The two oncogenes involved in these translocations are MTCP-1 and TCL-1, which constitute a family of genes involved in lymphoid proliferation and T cell malignancies (2, 5, 6).
The MTCP-1 (mature T cell proliferation-1) gene in the human X chromosome was the first candidate gene involved in the leukemogenesis of mature T cells (7). Aberrant or overexpressed MTCP-1 transcripts are found in the rare, but recurrent, chromosomal translocation t(X;14) in T cell proliferative diseases (8). About 10% of patients with the genetic disease Ataxia telangiectasia have clonal T cell proliferations with this cytogenetic aberration (1–3). MTCP-1 has two short ORFs that express A1 and B1 transcripts. The shorter A1 transcript is widely expressed in both tumor and nontumor cells. Expression of the B1 transcript is restricted to mature T cell proliferations with t(X;14) translocations (9), including T cell prolymphocytic leukemia tumors from patients with ataxia telangiectasia (10). The function of the B1 transcript of MTCP-1 is not known, but it encodes for a protein of 107 amino acids.
The TCL-1 oncogene maps at chromosome 14q32.1 (5). Chromosomal rearrangements at the TCL-1 locus are associated with T cell leukemia of the mature phenotype such as T cell prolymphocytic leukemia, adult T cell leukemia, and chronic T cell leukemias in patients with immunodeficiency syndrome Ataxia telangiectasia. TCL-1 is activated in T cell leukemias and lymphomas by chromosome translocations or inversions that juxtapose the α/δ or the β locus of the T cell receptor to the TCL-1 oncogene. TCL-1 is expressed at high levels in pre-B cells and immature thymocytes, but not in mature B or T cells (11). No expression of TCL-1 was observed in cell lines derived from normal human tissues. Ataxia telangiectasia patients with chromosome 14 rearrangements show expression of TCL-1 in leukemic T cells (10, 12). Similarly, the murine homolog of TCL-1 is expressed early in embryonic development in immature T and B cells (13).
The TCL-1 gene codes for a protein of 114 amino acid residues and molecular weight of 14,000 daltons (6). TCL-1 and MTCP-1 proteins share 40% identical amino acid residues and 61% similar amino acids, which strongly suggested that their tertiary structures are similar. However, no similarity was found with other human genes. Therefore, TCL-1 and MTCP-1 represent members of a unique family of genes involved in lymphoid proliferation and T cell malignancies. Human recombinant TCL-1 and MTCP-1 proteins have been expressed in Escherichia coli and purified for structural analysis to investigate their role in development of T cell malignancies (14). CD spectra at neutral pH are similar for both proteins and suggest β-sheet secondary structure. The crystal structure of MTCP-1 has been determined and refined to an R factor of 0.21 at 2.0 Å resolution. A model has been built of the structure of TCL-1. Analysis of these structures and the implications for the cellular function of these oncogene products are discussed.
MATERIALS AND METHODS
Purification and Crystallization of MTCP-1.
Human recombinant MTCP-1 protein was expressed and purified as described previously (14). Briefly, MTCP-1 protein was expressed in E. coli by using the prokaryotic expression vector pQE30 with a 6×His tag sequence followed by a thrombin cleavage site placed before the N terminus. The plasmid is designated pQE-H6-Thr-MTCP-1. The recombinant protein contains Gly-Ser at the amino terminus instead of Met. The protein was purified by metal chelate chromatography, digestion with thrombin, and reverse-phase FPLC chromatography. The purified protein was dialyzed into Tris buffer at pH 7.8 and concentrated to 5.0 mg/ml for crystallization. Crystals of MTCP-1 protein were grown by vapor diffusion at room temperature by using 1.5 M ammonium sulfate as precipitant. The crystals reached a size of 0.25 × 0.25 × 0.5 mm3 within a few days. X-ray diffraction data were collected on an R-AXIS II imaging plate detector mounted on an RU200 Rigaku rotating anode x-ray generator. X-ray diffraction data were measured to 2.0 Å resolution and reduced with the hkl package (HKL Research, Charlottesville, VA) (15). The native Fobs were further reduced by the trancate routine of Collaborative Computational Project 4 (16). The unit cell dimensions were a = b = 62.665 and c = 85.962 Å, and the space group was P6222, as determined by examination of the major zones for symmetry and the axes for systematic absences.
Purification and Crystallization of Selenomethionyl MTCP-1.
The expression plasmid pQE-H6-Thr-MTCP-1 was transformed into the B834 strain of bacteria, which is a methionine auxotroph (Novagen). The starter cultures were grown in M9 medium supplemented with 5% Luria–Bertani medium, and the cells were washed with M9 medium and grown in M9 methionine-deficient medium according to the procedure of Leahy et al. (17) using 5 μg/ml of Trp and Tyr, 50 μg/ml of the other 16 amino acids (no Met or Cys), and 50 μg/ml of selenomethionine. The Se-Met MTCP-1 protein was purified to homogeneity by using the same procedure as for native protein (14); however, all buffers were degassed and 1 mM β-mercaptoethanol was added to all buffer solutions. The yield of purified Se-Met MTCP-1 was about 10 mg per liter of culture. The Se-Met MTCP-1 was crystallized as described above for the native protein, with the addition of 1.0 mM 2-mercaptoethanol.
Determination of Crystal Structure.
Multiwavelength anomalous diffraction (MAD) data on a single Se-Met MTCP-1 crystal were collected at 90 K on the MAR300 image plate detector of beamline X12-B at National Synchrotron Light Source at Brookhaven National Laboratory, to obtain MAD phases (18). The crystal was transferred to an artificial mother liquor (2.5 M ammonium sulfate, 25% glycerol, 1.0 mM β-mercaptoethanol in Tris buffer at pH 7.8), and flash-cooled by putting into the nitrogen stream at 90 K. The diffraction data were processed with the hkl package. The unit cell dimensions were a = b = 62.549 and c = 85.278 Å in space group P6222. The positions of the three Se sites were determined with shelx (19) using Fa calculated by madsys (20). Initial phases were derived by madsys. The phases and figures of merit to 3.0 Å from madsys were used for density modification by routine dm of Collaborative Computational Project 4. The initial electron density map showed the large side chains and the entire polypeptide chain, except for three regions around Glu-5, Glu-19, and Thr-38. The initial model was built by using the program o (21) on a Silicon Graphics Indigo workstation. The tracing was confirmed after phase combination of the partial model, which consisted of residues 48–108, and polyalanine elsewhere, except for the above three regions, using the sigmaa routine of Collaborative Computational Project 4. The structure was refined by using native MTCP-1 data between 6.5 and 2.0 Å resolution with the program x-plor (22) and refitted with frodo (24). The working set and Rfree test set (23) for refinement contained 6,151 (91.1% complete) and 582 (8.6%) unique reflections, respectively. Omit maps were calculated for each two residues to confirm the structure. Twenty-four water molecules were located in Fo − Fc and 2Fo − Fc electron density maps. The final electron density map showed continuous density for the entire polypeptide chain, with the exception of residues 1 and 2, which are disordered. The topology was compared with other structures in the Protein Data Base (PDB) by using dejavu (G. T. Kleywegt and T. A. Jones, University of Uppsala, Sweden). The MTCP-1 coordinates are entry 1A1X in the PDB.
Molecular Model of TCL-1.
A similarity model was built for the related protein TCL-1. The initial sequence alignment was generated by maximizing the correlation between the sequences. The modeling program ammp (25) running on a 233 MHz PC in WindowsNT was used to build the model. The model was generated in two passes, first with an initial alignment based solely on sequence similarity and then with the position of the small insertion in the sequence shifted to provide a smaller deviation between the model and the starting coordinates. The realignment shifted the insertion from the end of a β strand to the first turn of the long surface loop. The model structure was built by energy minimization where short runs of conjugate gradients (100 cycles) were used with the SP4 version of the potential set and the SP4 potential set with harmonic restraints to the MTCP-1 structure alternately. The harmonic restraints are defined by K(r − r0)2 where K is the force constant and r is the x,y,z cartesian vector. An arbitrary force constant of 100 kcal/mol-Å2 was used. The final structure was minimized with the SP4 potential, which is a reparameterization of S and P atoms from the SP3 potential given in Weber and Harrison (26). The resulting model has a rms deviation between the TCL-1 model and MTCP-1 coordinates of 0.59 Å on all atoms, with a maximum deviation of 2.82 Å on the carbonyl oxygen of Arg-48, which is adjacent to the insertion. The TCL-1 model is highly similar to the MTCP-1 crystal structure and has no regions with high residual strain energy.
Calculation of Electrostatic Potential Surfaces.
Three-dimensional maps of the molecular electrostatic potential were calculated for the MTCP-1 crystal structure and the TCL-1 model. The electrostatic field (−Σ(q/r)) was calculated for each point on a 1 Å grid, which was sized so that the total map was 20 Å larger than the molecular dimensions (10 Å on each side). A constant dielectric of one was used, and the summations were performed with ammp using the partial charges in the SP4 potential set.
RESULTS
Crystal Structure of MTCP-1.
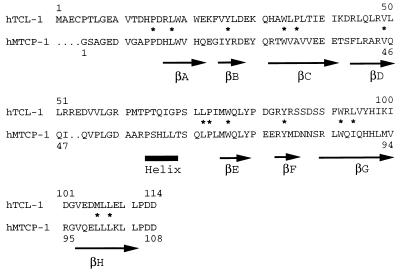
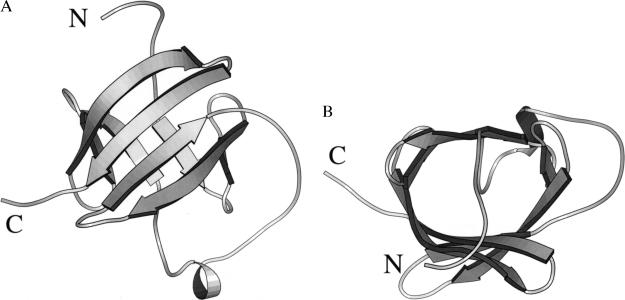
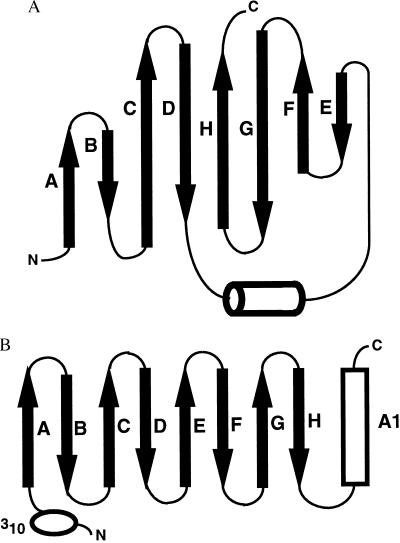
The crystal structure of recombinant human MTCP-1 was determined by multiwavelength anomalous diffraction phasing and refined to an R factor of 0.21 at 2.0 Å resolution with individual thermal factors and 24 water molecules. The statistics for data collection and refinement are shown in Table 1 and the Se sites in Table 2. The final 2Fo − Fc electron density map was continuous for all residues, except for the amino terminal two residues, which are disordered. The solvent content of 36.5% is relatively low because of the tight packing of protein molecules in this crystal lattice. procheck geometrical analysis (27) showed 85.9% of amino acid residues within the most favored regions of the Ramachandran plot and 14.1% in additional allowed regions. MTCP-1 folds into a compact eight-stranded antiparallel β barrel with a short helix between β strands 4 and 5 (Figs. 1 and 2). The secondary structure is consistent with CD spectra for both MTCP-1 and TCL-1, which showed predominantly β structure (14). The short helix in MTCP-1 consists of residues Pro-58 to Leu-62. The β barrel topology is illustrated in Fig. 3. The eight β strands form a continuous antiparallel sheet with the helix lying in the loop crossing over from strand D to E. The β barrel shows internal pseudosymmetry between the amino terminal four strands and the carboxy terminal four strands. This topology appears to be unique; no other example was found in a search of the Protein Data Base. One face of the β barrel is formed by the four longer strands C, D, G, and H, whereas the other four shorter strands form the opposite face. The internal hydrophobic core consists of residues Pro-11, Leu-14, Tyr-22, Trp-30, Ala-32, Val-45, Leu-66, Pro-67, Trp-70, Tyr-78, Trp-86, Ile-88, Leu-100, and Leu-102 (Fig. 1). The aromatic Trp-30, Trp-70, Tyr-78, and Trp-86 lie within the β barrel, whereas Tyr-22 lies between the β barrel and the helix. Both TCL-1 and MTCP-1 proteins show a relatively high Trp content that includes four conserved Trp residues. Trp is relatively rare in other proteins and may play an important role in the structure or function of these two proteins. The interior of the β barrel is completely filled with the amino acid side chains, and there is no empty space of significant size or unexplained electron density in the protein interior.
Table 1.
X-ray data collection and refinement statistics
| Dataset
|
|||||
|---|---|---|---|---|---|
| E1 | E2 | E3 | E4 | Native | |
| Wavelength Å | 0.98745 | 0.97980 | 0.97910 | 0.95741 | Cu Kα |
| Resolution Å | 28.4-2.2 | 28.4-2.2 | 28.4-2.2 | 28.4-2.2 | 21.1-2.0 |
| Rmerge | 0.046 | 0.050 | 0.061 | 0.055 | 0.047 |
| Reflections | |||||
| Observed | 36,489 | 37,220 | 36,626 | 37,901 | 42,682 |
| Unique | 5,339 | 5,402 | 5,395 | 5,418 | 7,213 |
| Completeness % | 98.1 | 99.7 | 99.7 | 99.9 | 99.7* |
| 〈I/ΣI〉 | 18.8 | 16.6 | 12.5 | 14.7 | 15.1 |
| f′† | −4.10 | −10.14 | −8.03 | −2.55 | |
| f"‡ | 0.51 | 3.45 | 5.37 | 3.93 | |
| Refinement statistics | |||||
| RMS deviation | |||||
| Resolution | Rcryst | Rfree | Bond length | Bond angle | Mean B factor |
| 6.5–2.0 Å | 0.211 | 0.253 | 0.007 Å | 1.4° | 21.4 Å3 |
The completeness in the 2.07–2.0 Å resolution shell was 99.3%.
The real part of the anomalous scattering factor of the selenium atom.
The imaginary part of the anomalous scattering factor of the selenium atom.
Table 2.
Location of selenium atoms
| Se sites | x | y | z | Occupancy |
|---|---|---|---|---|
| Se1 (Met-79) | 0.13162 | 0.22057 | 0.27665 | 1.02732 |
| Se2 (Met-69) | 0.07690 | 0.26150 | 0.20690 | 1.07054 |
| Se3 (Met-93) | 0.41234 | 0.65524 | 0.13024 | 0.98692 |
Figure 1.
Structural alignment of amino acid sequences of human recombinant MTCP-1 and TCL-1. The secondary structure of MTCP-1 is indicated below the sequence by arrows for β strands A to H and a thick line for the helix. The residues that form the internal hydrophobic core of the structure are indicated by ∗.
Figure 2.
Crystal structure of MTCP-1. A molscript (36) ribbon representation with arrows indicating β strands. Two orthogonal views are shown in A and B.
Figure 3.
(A) Topology of the eight-stranded antiparallel β barrel of MTCP-1. β strands A to H are indicated by arrows and the helix by a cylinder. (B) Topology of the lipocalin fold. The β strands A to H are indicated by arrows, the α helix by a cylinder, and the amino-terminal 310 helix by an ellipse.
Relationship of MTCP-1 to Lipocalin Family.
The crystal structure of MTCP-1 is superficially similar to the structures of the lipocalin family and the calycin superfamily of proteins. However, the topology of the β barrel is different (Fig. 3). The proteins in the calycin superfamily are distantly related in amino acid sequence, but share a common structural fold that consists of 8–10 antiparallel β strands that form a β barrel (28). The lipocalins are predominantly extracellular transport proteins that consist of about 150–183 residues in an eight-stranded β barrel that forms an internal binding site for a hydrophobic ligand. The structure includes an amino terminal 310 helix before the β barrel and a carboxy terminal α helix. Retinol binding protein is the best-characterized member of the lipocalin family (29). A second family within the calycin superfamily consists of mostly intracellular fatty acid binding proteins. These proteins fold into 10-stranded β barrels with an amino terminal 310 helix and two helices in the loop between the first two strands (30). Finally, the avidins fold into an amino terminal 310 helix followed by eight antiparallel β strands in the same arrangement as the lipocalin fold. MTCP-1 and TCL-1, with 107 and 114 residues, respectively, are significantly smaller than the typical lipocalin, and the β barrel has a different topology (Fig. 3). In the calycin superfamily, the β barrel has a simple up-down topology with each β strand lying between the two strands that are closest in the sequence in the order ABCDEFGH for the eight-stranded lipocalin family and the avidins, and ABCDEFGHIJ for the fatty acid binding proteins. In contrast, MTCP-1 has the strand order ABCDHGFE, with a short helix between strands D and E. These topological differences suggest that there is no common evolutionary origin. However, a simple rearrangement that translates the amino terminal four β strands to the carboxy terminus of the hypothetical protein can reproduce the lipocalin topology. Therefore, it is not clear if the overall similarity between the structures of MTCP-1 and lipocalins is the result of convergent evolution to a β barrel fold, or divergent evolution involving transposition of two halves of the protein. We propose that TCL-1 and MTCP-1 form members of a unique family of proteins because of the lack of sequence similarities, smaller size, and topological differences compared with the lipocalin family.
Molecular Model for TCL-1.
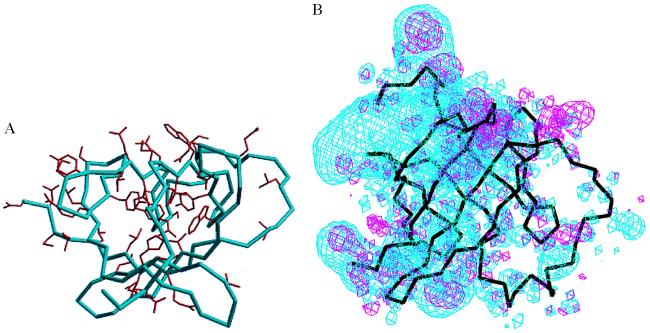
A molecular model was built for TCL-1 based on the 40% identity in amino acid sequence shared with MTCP-1 to analyze the structural similarities and differences and implications for their function. The sequence alignment deduced from the modeling is shown in Fig. 1. Human TCL-1 has four additional residues at the amino terminus compared with recombinant human MTCP-1 and has a two-residue insertion that was positioned in the surface loop between β strand D and the helix. The secondary and tertiary structures are predicted to be conserved in MTCP-1 and TCL-1, consistent with CD measurements (14). The 43 residues that are identical in TCL-1 and MTCP-1 are shown in Fig. 4A. These identical residues cluster within the center of the β barrel and on the face formed by the four shorter β strands. Most of the residues that form the hydrophobic core are identical in both proteins, including Pro-11, Leu-14, Tyr-22, Trp-30, Leu-66, Pro-67, Trp-70, Tyr-78, Trp-86, and Leu-102 (Fig. 1). Only four of these internal residues differ in MTCP-1 and TCL-1: Ala-32 is Pro in TCL-1, Val-45 is Leu, Ile-88 is Leu, and Leu-100 is Met in TCL-1. These changes all are conservative substitutions, except for the Ala/Pro 32, however, they will tend to increase the size of the internal hydrophobic residues. The clustering of conserved residues on one face of the β barrel suggests that the two proteins bind similar molecules at this surface.
Figure 4.
(A) The amino acid side chains of residues that are identical in TCL-1 and MTCP-1 are shown in red on the Cα backbone of the MTCP-1 structure (blue) looking into the β barrel in the same orientation as Fig. 2B. Electrostatic potential surface for MTCP-1 and TCL-1. The negative electrostatic potential energy surface contoured at 0.3 e/Å3 is shown in blue for the crystal structure of MTCP-1 and in red for the model of TCL-1. The black lines indicate the Cα backbone of MTCP-1 in the same view as Fig. 2A, showing the four longer strands crossing over the four shorter strands of the β barrel with the helix at the lower right side.
The electrostatic potential surfaces were calculated for the crystal structure of MTCP-1 and the model of TCL-1 (Fig. 4B). The negative electrostatic potential surfaces were distinctly different for the two proteins. MTCP-1 has a high distribution of negative charge around the face of the β barrel that is farthest from the helix. In contrast, TCL-1 did not show such a distinctive charge distribution. The negative electrostatic distribution of MTCP-1 lies close to Glu 5–6, Glu 34–36, Glu 75–76, and Asp 107–108. This asymmetric charge distribution suggests that the negatively charged surface of MTCP-1 may bind a positively charged region of a receptor, or that a hydrophobic molecule may bind on the opposite side near the helix of MTCP-1. In contrast, TCL-1 tends to be more hydrophobic or neutral on the surface.
DISCUSSION
The crystal structure of MTCP-1 has been determined at high resolution and used to predict the structure of TCL-1. Despite the detailed structural information, the role of these oncogene products in the development of T cell malignancies is not obvious. The tertiary structure of MTCP-1 and TCL-1 showed that these proteins are not canonical members of the lipocalin family, although they share a similar shape and eight-stranded antiparallel β barrel. However, it is worthwhile to consider the biological functions of the lipocalins to obtain clues to the cellular function of TCL-1 and MTCP-1. The lipocalin family is functionally diverse with roles in retinol transport, olfaction, sterol and pheromone transport, invertebrate coloration, prostaglandin synthesis, immune modulation, and cell regulation (28). The family is defined by identical tertiary folds in several crystal structures of different members, despite a rather low level of sequence identity of about 20% or less between pairs of the proteins (31). In addition, the lipocalins of known function are extracellular proteins that bind small hydrophobic ligands, including retinol, biliverdin, steroids, and prostaglandin. The superficial structural similarity between MTCP-1 and the lipocalin family of proteins suggests that MTCP-1 and TCL-1 also may bind small hydrophobic ligands within the β barrel structure, as in retinol binding protein. Alternatively, the structural similarities with the intracellular fatty acid binding proteins suggest that small hydrophobic ligands may bind between the β barrel and the helix of MTCP-1. Lipocalins, such as retinol binding protein, also bind other proteins. Retinol binding protein usually is complexed with transthyretin in plasma, and the affinity for transthyretin is higher when retinol is bound than for the apo protein. Retinol binding protein also binds a specific cell surface receptor that may be involved in the transfer of retinol to the cytoplasm. The implications for the cellular function of MTCP-1 and TCL-1 are that they are likely to be transport proteins for small hydrophobic ligands, although other lipocalins act as enzymes (prostaglandin D synthase) or protease inhibitors (32). Some members of the lipocalin family are involved in cell regulation and cancer. A retina lipocalin, purpurin, binds retinol and glycosaminoglycan and is believed to function in the control of cell differentiation, and survival (33). Secretion of apolipoprotein D is correlated with inhibition of proliferation of certain human breast cancer cell lines (34). Overexpression of neu-related lipocalin is observed in neu mammary carcinomas (35). Therefore, structural similarities with lipocalins are consistent with a similar role of TCL-1 and MTCP-1 in control of T cell differentiation and survival. In addition, murine TCL-1 is expressed in fetal hematopoietic organs and immature T and B cells early in embryonic development (13). MTCP-1 and TCL-1 are postulated to act by binding a small hydrophobic ligand that promotes a conformational change in the protein and increases its affinity for a receptor protein. Increased affinity of TCL-1 or MTCP-1 for the receptor protein is expected to increase T cell survival. Experiments to test this hypothesis are in progress.
Acknowledgments
We thank Charles Reed, Bhuvaneshwari Mahalingam, Vinod Kumar, and Malcolm Capel for help with synchrotron data collection. Charles Reed also helped prepare Figs. 2 and 4. We thank Wayne Hendrickson for the madsys program. Multiwavelength anomalous diffraction data were collected at the National Synchrotron Light Source, Brookhaven National Laboratory. This research was funded in part by the Elsa U. Pardee Foundation (I.T.W. and R.W.H.). Z.Q.F. is a Cure for Lymphoma Foundation Fellow.
Footnotes
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, Department of Biology, Brookhaven National Laboratory, Upton, NY 11973 (reference no. 1A1X).
References
- 1.Taylor A M R. In: Ataxia telangiectasia: A Cellular and Molecular Link Between Cancer, Neuropathology and Immune Deficiency. Bridges B A, Harnden D G, editors. New York: Wiley; 1982. pp. 53–81. [Google Scholar]
- 2.Stern M H. In: The Causes and Consequences of Chromosomal Aberrations. Kirsch I R, editor. Boca Raton, FL: CRC; 1993. pp. 165–176. [Google Scholar]
- 3.Stern M H, Zhang F, Griscelli C, Thomas G, Aurias A. Hum Genet. 1988;78:33–36. doi: 10.1007/BF00291230. [DOI] [PubMed] [Google Scholar]
- 4.Croce C M. Cell. 1987;49:155–156. doi: 10.1016/0092-8674(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 5.Virgilio L, Isobe M, Narducci M G, Carotenuto P, Camerini B, Kurosawa N, Abbas-ar-Rushdi, Croce C M, Russo G. Proc Natl Acad Sci USA. 1993;90:9275–9279. doi: 10.1073/pnas.90.20.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu T, Virgilio L, Narducci M G, Facchiano A, Russo G, Croce C M. Cancer Res. 1994;54:6297–6301. [PubMed] [Google Scholar]
- 7.Stern M H, Soulier J, Rosenzwajg M, Nakahara K, Canki-Klain N, Aurias A, Sigaux F, Kirsch I R. Oncogene. 1993;8:2475–2483. [PubMed] [Google Scholar]
- 8.Soulier J, Madani A, Cacheux V, Rosenzwajg M, Sigaux F, Stern M H. Oncogene. 1994;9:3565–3570. [PubMed] [Google Scholar]
- 9.Madani A, Soulier J, Schmid M, Plichtova R, Lerme F, Gateau-Roesch O, Garnier J P, Pla M, Sigaux F, Stern M H. Oncogene. 1995;10:2259–2262. [PubMed] [Google Scholar]
- 10.Thick J, Metcalf J A, Mak Y F, Beatty D, Minegishi M, Dyer M J S, Lucas G, Taylor A M R. Oncogene. 1996;12:379–386. [PubMed] [Google Scholar]
- 11.Virgilio L, Narducci M G, Isobe M, Billips L G, Cooper M D, Croce C M, Russo G. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narducci M G, Virgilio L, Isobe M, Stoppacciaro A, Elli R, Fiorilli M, Carbonari M, Antonelli A, Chessa L, Croce C M, et al. Blood. 1995;86:2358–2364. [PubMed] [Google Scholar]
- 13.Narducci M G, Virgilio L, Engiles J B, Buchberg A M, Billips L, Facchiano A, Croce C M, Russo G, Rothstein J L. Oncogene. 1997;15:919–926. doi: 10.1038/sj.onc.1201246. [DOI] [PubMed] [Google Scholar]
- 14.Du Bois, G. C., Song, S. P., Kulikovskaya, I., Virgilio, L., Varnum, J., Germann, M. W. & Croce, C. M. (1997) Protein Expression Purif., in press. [DOI] [PubMed]
- 15.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 17.Leahy D J, Erickson H P, Aukhil I, Joshi P, Hendrickson W A. Proteins. 1994;19:48–54. doi: 10.1002/prot.340190107. [DOI] [PubMed] [Google Scholar]
- 18.Leahy D J, Hendrickson W A, Aukhil I, Erickson H P. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 19.Sheldrick G M, Dauter Z, Wilson K S, Hope H, Sieker L C. Acta Crystallogr D. 1993;49:18–23. doi: 10.1107/S0907444992007364. [DOI] [PubMed] [Google Scholar]
- 20.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 21.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brunger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 23.Brunger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Jones T A. Appl Crystallogr. 1978;11:268–272. [Google Scholar]
- 25.Harrison R W, Chatterjee D, Weber I T. Proteins. 1995;23:463–471. doi: 10.1002/prot.340230402. [DOI] [PubMed] [Google Scholar]
- 26.Weber I T, Harrison R W. Protein Sci. 1997;6:2365–2374. doi: 10.1002/pro.5560061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Flower D R. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowan S W, Newcomer M E, Jones T A. Proteins. 1990;8:44–61. doi: 10.1002/prot.340080108. [DOI] [PubMed] [Google Scholar]
- 30.Zanotti G, Scapin S, Spadon P, Veerkamp J H, Sacchettini J C. J Biol Chem. 1992;267:18541–18550. doi: 10.2210/pdb2hmb/pdb. [DOI] [PubMed] [Google Scholar]
- 31.Sansom C E, North A C T, Sawyer L. Biochem Biophys Acta. 1994;1208:247–255. doi: 10.1016/0167-4838(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 32.van’t Hof W, Blankenvoorde M F, Veerman E C I, Amerongen A V. J Biol Chem. 1997;272:1837–1841. doi: 10.1074/jbc.272.3.1837. [DOI] [PubMed] [Google Scholar]
- 33.Berman P, Gray P, Chen E, Keyser K, Erlich D, Karten H, LaCorbiere M, Esch F, Schubert D. Cell. 1987;51:135–142. doi: 10.1016/0092-8674(87)90018-3. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Boado Y-S, Toliva J, Lopez-Otin C. J Biol Chem. 1994;269:26871–26878. [PubMed] [Google Scholar]
- 35.Stoesz S P, Gould M N. Oncogene. 1995;11:2233–2241. [PubMed] [Google Scholar]
- 36.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]