Abstract
Several important and possibly interrelated functions have been identified for the HIV-1 accessory gene product Vpr. These include import of the HIV reverse transcription complex into the nucleus of nondividing cells, cellular differentiation including cell cycle arrest at the G2/M phase border, immune suppression, and enhancement of virus replication. We have cloned a candidate Vpr ligand, termed human Vpr interacting protein (hVIP/MOV34), by using a yeast two-hybrid assay. This gene is homologous to a simultaneously identified 34-kDa human mov34 homologue. The MOV34 family includes proteins that function as transcriptional and proteolytic regulators of cell growth and differentiation. We demonstrate direct interactions between the putative ligand hVIP/MOV34 and Vpr in vitro and in vivo. hVIP/MOV34 localizes to the nucleus and appears to function as a component of the cell cycle cascade. We observe an association between the induction of cell cycle arrest at the G2/M phase border by Vpr and a change in the subcellular localization of hVIP/MOV34 from a nuclear to a perinuclear localization. This was further associated with the inhibition of maturation promoting factor-associated histone H1 kinase activity. We conclude that hVIP/MOV34 is involved in the regulation of the cell cycle and a likely cellular cofactor for HIV-1 Vpr.
The HIV-1 accessory gene vpr, although dispensable for virus replication in T cell lines and activated primary peripheral blood mononuclear cells (PBMCs) (1–5), is required for efficient viral replication in primary monocyte/macrophages. HIV-1 vpr encodes a 14-kDa protein expressed primarily from a singly spliced Rev-dependent mRNA (3, 6–9). Vpr can modestly transactivate the HIV-1 long terminal repeat (3) and thus may up-regulate viral gene expression in newly infected cells before the appearance of Tat. It has been found to enhance the nuclear migration of the preintegration complex in newly infected nondividing cells (10). Significantly, Vpr induces cellular differentiation, which includes the activation of specific host cell gene transcription and growth arrest in primary cells and several tumor cell lines, even in the absence of any other viral proteins (11–14). Vpr blocks cell cycling at G2/M phase of the cell cycle (15). This finding has been associated with a change in the phosphorylation state of CDC2 kinase (16, 17) and may involve in direct regulation of host transcription and modulation of immune responsiveness (12). Furthermore, Vpr expression appears to inhibit the establishment of chronic infection in cells (15, 18, 19). Macreadie et al. (20) reported that Vpr causes growth arrest and structural defects in yeast. Functional studies have shown that Vpr accelerates HIV-1 replication in some T lymphoid cell lines and in primary macrophages particularly early in infection where the effects of Vpr are more pronounced (21–23).
Mutational analysis of the 96-amino acid Vpr protein has identified distinct functional domains. An amino-terminal acidic putative α-helix acts as a nuclear localization signal and is essential for the virion incorporation of Vpr through interaction with Gag (17). Toward the carboxyl terminus of Vpr lies a short leucine/isoleucine-rich sequences termed the LR domain and a HFRIGCRHSRIG motif that is dispensable for virion incorporation but, nevertheless, important for nuclear localization and cell cycle arrest activity of Vpr (17, 20). Although no classical nuclear localization signal has been clearly identified for Vpr, it has been suggested that Vpr may gain access to the nucleus by specific interactions with proteins that migrate from cytoplasmic to nuclear locations (24, 25). Recent studies have suggested that the LR domain is a Vpr cofactor binding domain because Vpr protein mutated in the LR domain has a cytoplasmic localization and an inability to arrest cells at G2/M phase of cell cycle (17, 25). If we assume that Vpr functions directly during cell cycle arrest and nuclear transport of the HIV-1 preintegration complex into the nucleus of the nondividing cells, the identification of cellular factors contacted by Vpr might illuminate cellular strategies used for Vpr function and also generate insight into cell cycle transition by a wide variety of cells. Herein, we report the identification and initial characterization of a cellular factor that interacts with HIV-1 Vpr. This factor termed human Vpr interacting protein (hVIP/MOV34) is identical to the human 34-kDa mov34 homologue, a protein belonging to a gene family of transcriptional regulators, proteosome family members, and proteins involved in the regulation of the cell cycle.
MATERIALS AND METHODS
Yeast Two-Hybrid Interaction Assay.
Full-length HIV-1 Vpr was fused in-frame with the GAL-4 DNA binding domain in the yeast expression vector pGBT9 (CLONTECH). The two-hybrid screen was performed with a GAL-4 activation domain-tagged PBL cDNA library (CLONTECH) and a GAL-4 Vpr construct that was used as bait. After 4 days of selection on culture plates, double transformants were transferred to filter paper (VWR) and analyzed for β-galactosidase expression according to the manufacturer’s protocol (CLONTECH). Three clones were recovered and all contained the carboxyl-terminal portion of hVIP/MOV34 starting at amino acid 31. 5′ rapid amplification of cDNA ends was performed to isolate the 5′ end of the hVIP/MOV34 gene fragment by using total RNA from CEM and HeLa cells according to the instruction manual (Boehringer Mannheim). All the molecular clones were sequenced with the dideoxynucleotide chain-termination method (United States Biochemical) and the automatic sequencing method using Amplitag (Perkin–Elmer).
BLAST Search and Secondary Structure Analysis.
The amino acid sequence was analyzed against the known sequences deposited in the GenBank with blast search and the search was performed at the National Center for Biotechnology Information with the GENINFO(R) Experimental BLAST Network Service (26). Secondary structure was calculated by the nearest neighbor segment method of Solovyev and Salamov described at the Worldwide Web site at the Baylor College of Medicine secondary structure prediction home page.
RNA and Protein Expression.
Multitissue Northern blots were purchased from CLONTECH, and total RNA was isolated from CEM and RD cells and PBMCs. The blots were probed with a random-primed [32P]dCTP-labeled probe prepared from an internal 1,009-bp EcoRI hVIP/MOV34 cDNA fragment. A T7 RNA polymerase (vFT7–3) based expression system was used to assay the expression of hVIP/MOV34 in tissue culture cells (27, 28). hVIP/MOV34 cDNA was fused in-frame with an anti-Xpress antibody epitope tagged six-His mammalian expression vector pcDNA3.His (Invitrogen) and used for expression studies. HeLa cells were infected with vFT7–3 and transfected with the hVIP expression plasmid by using Lipofectin (GIBCO/BRL). After overnight transfection, the cells were labeled with 35S Express protein labeling mixture (NEN) for 2 h, and hVIP/MOV34 was immunoprecipitated with anti-Xpress mAb and subjected to SDS/PAGE in 15% gels.
Coimmunoprecipitation Assay.
Equal levels of in vitro translated hVIP/MOV34, HIV-1 Vpr, and p55 Gag precursor polyprotein were mixed and incubated for 30 min at 4°C in a binding buffer containing 25 mM Hepes (pH 7.9), 150 mM KCl, 0.1% Nonidet P-40, 5% glycerol, 0.5 mM DTT, and 0.4 mM phenylmethylsulfonyl fluoride. Respective antibodies were added to each tube with 150 μl of binding buffer and incubated for 90 min at 4°C. Protein A-Sepharose (5 mg per tube) was added to all the tubes, which were then incubated at 4°C for 90 min in a rotating shaker. The beads were then washed three times with the binding buffer. The immunoprecipitated protein complexes were eluted from the Sepharose beads and subjected to SDS/PAGE in 15% gels. The gel was processed for fluorography as described (17).
Cell Cycle Analysis.
HeLa or RD cells were cotransfected with either wild-type or mutant Vpr and hVIP/MOV34 sense or antisense expression vectors with pBabepuro (a vector that expresses the puromycin gene). Two days later, puromycin was added at a concentration of 2 μg/ml to eliminate the untransfected cells. The transfected cells were stained with propidium iodide 5–7 days after transfection for analysis of DNA content by flow cytometry.
Indirect Immunofluorescence.
HeLa cells were transfected with either Vpr or hVIP/MOV34 expression vectors alone or in combination. Immunofluorescence staining of fixed HeLa cells with the indicated primary antibody followed by fluorescein isothiocyanate (FITC; Boehringer Mannheim)- or rhodamine (Sigma)-conjugated secondary antibody was performed as described (28). Indirect immunofluorescence was carried out with rabbit anti-Vpr (1:50 dilution) or mouse anti-Xpress mAb (1:100 dilution; Invitrogen) alone or in combination. This was followed by staining with either rhodamine-conjugated goat anti-rabbit IgG (1:75 dilution) or FITC-conjugated goat anti-mouse IgG (1:100 dilution) or both.
Histone H1 Kinase Assay.
Vpr and hVIP/MOV34 sense and antisense expression vectors were used to transfect RD cell and the extracts were prepared by resuspending the cells in ice-cold HB buffer [25 mM Mops, pH 7.2/60 mM β-glycerophosphate/15 mM p-nitrophenyl phosphate/15 mM MgCl2/15 mM EGTA/1 mM DTT/0.1 mM sodium vanadate/1% Triton X-100/1 mM phenylmethylsulfonyl fluoride/leupeptin (20 μg/ml)/aprotonin (40 μg/ml)]. Lysates were repeatedly passed through a 26-gauge needle. Immunoprecipitation of cyclin B1-specific histone H1 kinase was performed with anti-human cyclin B1 antibody (Upstate Biotechnology) conjugated to protein A-agarose beads as described (29). For kinase activity determination, the samples were incubated with histone H1 (Boehringer Mannheim) in the presence of [γ-32P]ATP for 20 min. The reaction was stopped by the addition of 2× sample buffer and boiled for 5 min, and fractionation was performed by SDS/PAGE on 12% gels. Gels were dried prior to autoradiography, and densitometry was performed by using nih image 1.61 software.
RESULTS
By using a yeast two-hybrid protein interaction trap, we sought to identify a human cDNA encoding a protein able interact with Vpr (30). We isolated several potential ligands; however, three clones fulfilled criteria essential for a Vpr ligand, containing overlapping complementary DNA derived from the same gene. 5′ rapid amplification of cDNA ends was used to isolate a full-length 1.35-kb cDNA containing a 1,023-bp ORF. The protein encoded by this cDNA was designated hVIP. Comparison of the hVIP cDNA sequence with the data bank yielded an almost perfect match starting at amino acid 45 of the hVIP sequence with amino acid 1 of a simultaneously identified 34-kDa human Mov34 homologue (ref. 31; deposited after submission of this manuscript). The human 34-kDa MOV34 homologue represents a protein of unknown function but a member of the MOV34 gene family (Fig. 1a). This family includes several subset members of the human eukaryotic initiation factor 3 complex (eIF3), subunits of the 26S proteasome S12 complex, and several related transcriptional factors (32). These proteins function as multimeric complexes involved in regulation of host cell transcription, cell division, cell fate determination, and RNA functions (31, 32). To place hVIP in the MOV34 family, we have adopted the name as hVIP/MOV34. Nine dipeptide leucine/isoleucine motifs and several leucine/isoleucine repeats were noted in the amino acid sequence of hVIP/MOV34. Such leucine/isoleucine repeats are believed to mediate protein–protein interactions (33, 34). The leucine/isoleucine repeats are found in predicted β-strands (positions 12–17, 61–63, 111–115, 165–169, and 200–205), α-helical regions (positions 103–104, 189–190, 236–237, 251–253, and 271–272), and coil regions. Initial predictions of exposed regions associated with the leucine/isoleucine regions indicate that the segments around positions 100–140, 200–220, and 240–260 are predominately exposed based upon theoretical considerations of solvent accessibility (data not shown). These regions correspond to both predicted β-strand and α-helical domains (Fig. 1a). Although the homologue belongs to the Mov34 family, comparison of secondary structure and solvent accessibility profiles of eIF3 WD-repeat family members suggest that the proteins display exposed regions involving predominately β-strand structures but display no obvious sequence similarity with hVIP/MOV34 (unpublished observations).
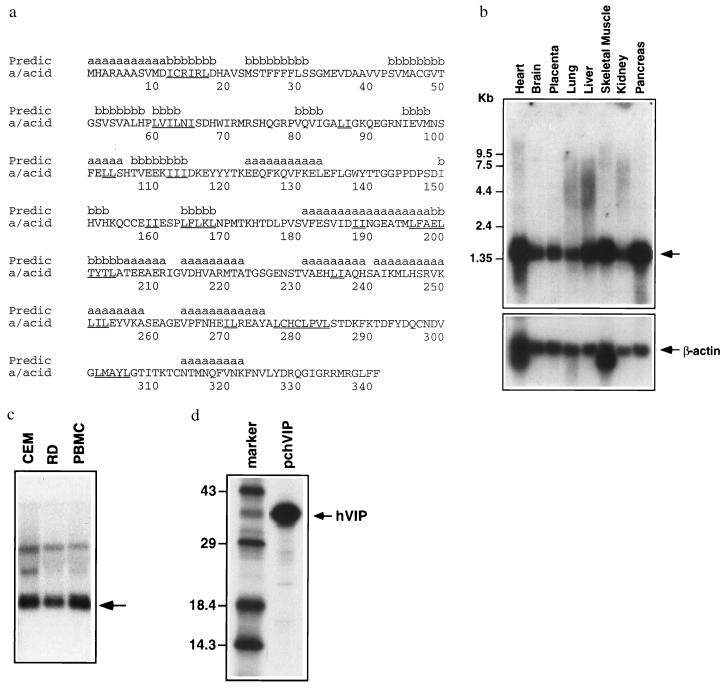
Figure 1.
Identification and characterization of hVIP/MOV34. (a) Amino acid sequence of hVIP/MOV34. Secondary structure prediction is shown above the sequence. Leucine (L) or Isoleucine (I) repeats are underlined in the sequence. Northern blot of poly(A)+ RNA from a variety of human organs (b) and RNA from CEM and RD cells and PBMCs (c) were probed with a 32P-labeled hVIP/MOV34 fragment. (d) hVIP/MOV34 expression was detected by an immunoprecipitation assay. hVIP/MOV34 cDNA was fused in-frame with an anti-Xpress epitope tagged with a six-His mammalian expression vector pcDNA3.His (Invitrogen). HeLa cells were transfected with hVIP/MOV34 expression vectors and then labeled with a 35S protein labeling mixture (NEN), and hVIP/MOV34 was immunoprecipitated with anti-Xpress antibody and analyzed by SDS/PAGE on 15% gels.
The hVIP/MOV34 cDNA is approximately 1.35 kb, which corresponds to the size detected in Northern blots containing poly(A)+ RNA from several human organs and RNA from CEM cells, PBMCs, and a human rhabdomyosarcoma cell line (RD) with hVIP/MOV34 cDNA as a probe (Fig. 1 b and c). This diverse expression pattern suggests a role for hVIP/MOV34 in the biology of divergent cell lines not merely as cellular targets for HIV-1. Indeed, the murine homologue of the MOV34 gene is essential for embryonic development (35).
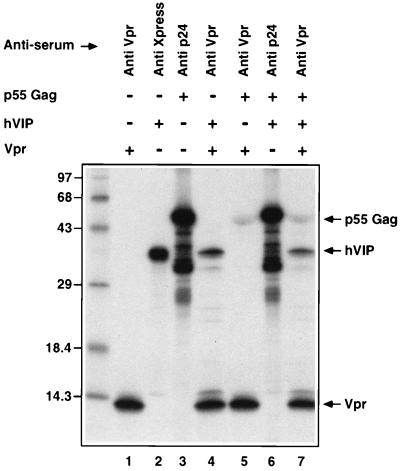
The biology of hVIP/MOV34 was investigated by fusing full-length hVIP cDNA in-frame with an anti-Xpress epitope tagged six-His mammalian expression vector, pcDNA3.His. hVIP/MOV34 cDNA expresses a polypeptide of relative molecular mass of 39,000 (Mr = 39,000), which was detected by immunoprecipitation with anti-Xpress epitope mAb (Fig. 1d). In an effort to further demonstrate the existence of a physical interaction between Vpr and hVIP/MOV34 as suggested by the yeast two-hybrid assay, we investigated whether Vpr would form a complex with hVIP/MOV34 in vitro. For this purpose, we mixed equal amounts of 35S-labeled in vitro-translated hVIP/MOV34 with 35S-labeled Vpr and/or HIV-1 p55 gag precursor polyprotein (as control). The relevant mixtures were incubated with their respective antiserum and their interaction was analyzed by coimmunoprecipitation, followed by SDS/PAGE. Fig. 2 shows that both Vpr and hVIP/MOV34 were coimmunoprecipitated by anti-Vpr antiserum. Polyclonal antiserum against p24 gag did not immunoprecipitate hVIP/MOV34. Interestingly, anti-Vpr antiserum immunoprecipitated both p55 gag and hVIP/MOV34, suggesting a lack of competition and distinct binding sites on Vpr for both ligands. Such a finding would be supported that Vpr as part of the viral preintegration complex could still interact with hVIP/MOV34. These data demonstrate that the specific interaction of hVIP/MOV34 and Vpr can be recapitulated in vitro and that the interaction is direct.
Figure 2.
Interaction of hVIP/MOV34 with HIV-1 Vpr in vitro by a coimmunoprecipitation assay. Equal amounts of 35S-radiolabeled in vitro-translated hVIP/MOV34 was incubated with HIV-1 Vpr and p55 gag precursor polyprotein on ice with binding buffer for 30 min. The protein complexes were immunoprecipitated with anti-Vpr, anti-Xpress, or anti-p24 antibodies and then subjected to SDS/PAGE on 15% gels. The hVIP/MOV34 protein did not coimmunoprecipitate with p55 gag and anti-p24 antibody but did coimmunoprecipitate specifically with Vpr and anti-Vpr antiserum. The relative mobility of the marker protein is indicated at the left in kilodaltons.
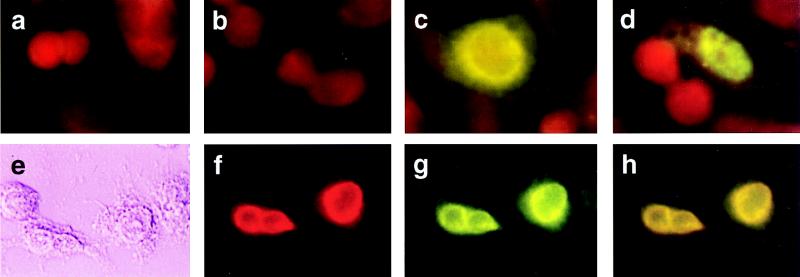
We next looked to see whether hVIP/MOV34 and Vpr staining would colocalize to the nucleus of cotransfected cells. When expressed independently, virtually all of the hVIP/MOV34 was present in the nucleus forming a punctate staining pattern (Fig. 3d), whereas Vpr is localized to the periphery of the nucleus (Fig. 3c). Two-color immunostaining with rabbit polyclonal Vpr antibody and anti-Xpress epitope mAb revealed that the coexpression of Vpr and hVIP/MOV34 strongly altered the distribution of hVIP/MOV34 from a punctate nuclear to a perinuclear pattern (Fig. 3g). Significant colocalization of hVIP/MOV34 and Vpr was observed in the periphery of the nucleus in the presence of Vpr (Fig. 3h). The alteration in subcellular localization of hVIP/MOV34 induced by coexpression with wild-type Vpr was specific in that it was not altered by cotransfection with an expression vector encoding mutant Vpr molecules (Fig. 4). This suggests that HIV-1 Vpr interacts with hVIP/MOV34 and alters the subcellular distribution of hVIP and supports the assertion that hVIP/MOV34 and Vpr physically interact in vivo.
Figure 3.
Subcellular distribution and colocalization of hVIP/MOV34 and Vpr by indirect immunofluorescence assay. (a–d) HeLa cells were transfected with either hVIP/MOV34 or HIV-1 Vpr expression vectors, fixed for immunofluorescence as described previously (28), probed with anti-Vpr (a and c) and anti-Xpress antibody (b and d), and stained with fluorescein-conjugated goat anti-rabbit for Vpr or goat anti-mouse secondary antibody for hVIP. hVIP localized to the nucleus with a punctate staining pattern and Vpr localized to the periphery of the nucleus. a and b were vector-transfected; c, Vpr-transfected; d, hVIP/MOV34-transfected. (e–h) Colocalization of hVIP/MOV34 and Vpr. HeLa cells were cotransfected with hVIP/MOV34 and HIV-1 Vpr expression vectors and then fixed for immunofluorescence. Fixed cells were probed with anti-rabbit polyclonal primary antibody followed by staining with rhodamine-conjugated goat anti-rabbit secondary antibody for Vpr. The cells were probed with anti-Xpress mAb, followed by fluorescein-conjugated goat anti-mouse secondary antibody for hVIP/MOV34. (e) Phase-contrast field. (f) Rhodamine-specific Vpr fluorescence. (g) Fluorescein-specific hVIP/MOV34 fluorescence. (h) Double exposure in which the mixture of rhodamine and fluorescein appears yellow.
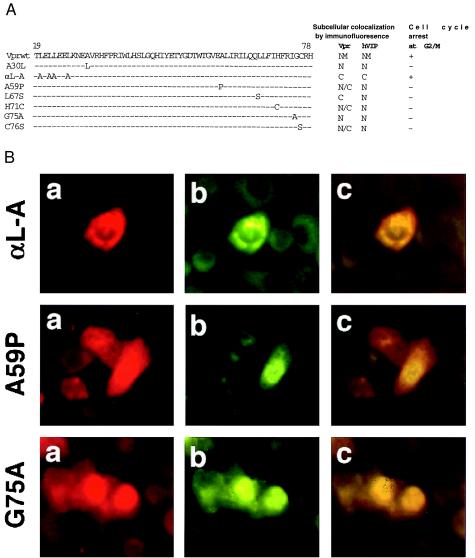
Figure 4.
Colocalization and interaction of hVIP/MOV34 with HIV-1 Vpr mutants. (a) Mutant Vpr molecules were generated by overlap PCR as described (17, 28). HeLa cells were cotransfected with hVIP/MOV34 and various Vpr mutants (A30L, αL-A, A59P, L67S, H71C, G75A, and C76S). The transfected cells were fixed and stained as described above and visualized with fluorescence and rhodamine wavelength filters to detect expression, and the cell cycle arrest activity of Vpr mutants was determined by flow cytometric analysis. (b) Subcellular distribution of hVIP/MOV34 in various Vpr mutants was assayed by indirect immunofluorescence.
To identify the domains of Vpr required for its interaction with hVIP/MOV34 and cell cycle arrest, we generated various mutant Vpr molecules by overlap PCR as described (28). The correlation between the localization of Vpr mutants with hVIP/MOV34 and cell cycle arrest was assessed. We cotransfected HeLa cells with expression vectors for different Vpr mutants and the hVIP/MOV34 expression plasmids and studied their subcellular localization. Fig. 4 shows that Vpr mutants A30L, A59P, L67S, H71C, G75A, and C76S do not alter the subcellular distribution of hVIP/MOV34 (Fig. 4 a and b) nor inhibit cellular proliferation (Fig. 4b), whereas hVIP/MOV34 localized to the cytoplasm when coexpressed with Vpr mutant αL-A (Fig. 4a). This mutant maintains the cell cycle arrest function of wild-type Vpr and prevents the nuclear import of hVIP/MOV34 (Fig. 4b). These data clearly demonstrate that the amino acid residues in the carboxyl terminus of Vpr are important for the interaction between Vpr and hVIP/MOV34 and for altering the subcellular localization of hVIP/MOV34 in cell cycle arrest.
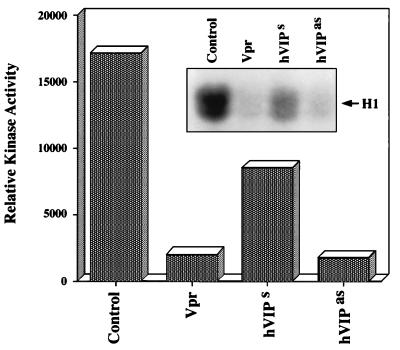
To confirm that hVIP/MOV34 is essential for the transition from G2 to M phase of cell division, we constructed an antisense hVIP/MOV34 expression vector. RD cells were transfected with Vpr and hVIP/MOV34 sense and antisense expression vectors and were selected with puromycin. Fig. 5 a and b reveal a significant reduction in the number of cells in Vpr- and hVIP/MOV34 antisense-transfected plates (Fig. 5a) and further show that the cells expressing Vpr and antisense hVIP/MOV34 were blocked at the G2/M phase of the cell cycle (Fig. 5b). Transition of G2 to M phase and induction of mitosis in cycling cells depends on the activation of maturation promoting factor (MPF) (36, 37), a protein kinase complex composed of a catalytic subunit, p34cdc2 (38–40), and its essential regulatory subunit, cyclin B (37). In cycling cells, MPF activity is undetectable during interphase but rises at the G2/M transition to induce mitosis. To explore whether hVIP/MOV34 has a role in regulating MPF associated kinase activity, we measured kinase activity in cells that were transfected with Vpr, hVIP/MOV34 sense, or antisense expression vectors. Kinase activity was inhibited by 90% in cells expressing Vpr and hVIP/MOV34 antisense compared with control cells (Fig. 6). These results clearly indicate that hVIP/MOV34 affects the activity of MPF and that the observed G2 arrest by Vpr may be due to the inhibition of kinase activity, which is essential for the cells to enter into mitosis.
Figure 5.
Antisense hVIP/MOV34 inhibits cellular proliferation. (a) Human embryonal rhabdomyosarcoma (RD) cells were transfected with Vpr, hVIP/MOV34 sense, and antisense expression vectors, and the cells were maintained in DMEM containing puromycin (2 μg/ml). The cells were photographed 5–7 days later with a Nikon phase-contrast microscope. (a) Control vector pBabepuro. (b) Vpr. (c) hVIP sense. (d) hVIP/MOV34 antisense. (b) The transfected RD cells were stained with propidium iodide and used for flow cytometric analysis as described (14). RD cells expressing Vpr and hVIP/MOV34 antisense arrest at G2/M phase of the cell cycle with 4n DNA content.
Figure 6.
Effect of Vpr, hVIP/MOV34 sense, and hVIP/MOV34 antisense expression on histone H1 kinase activity. Cyclin B1 was immunoprecipitated from RD cell extracts and incubated with [γ-32P]ATP in the presence of exogenously added histone H1. Quantification of kinase activity by densitometry scanning was plotted. (Inset) Autoradiogram.
DISCUSSION
The Vpr gene product encoded by HIV-1 is of considerable scientific interest. Vpr function is critical for the transport of HIV-1 preintegration complex into the nucleus of nondividing cells, and it increases virus replication in monocyte and primary cells (11, 23). Vpr is a retroviral accessory protein that modulates host cell function by arresting the cells at G2/M phase of the cell cycle (14, 15). Recently, it been shown that Vpr regulates apoptosis in target cells through suppression of NF-kB activity via the induction of IkB, supporting a role for HIV-1 Vpr in modulation of the host immune responses (12). Vpr therefore represents an attractive target for chemotherapeutic intervention of HIV-1-induced disease. Such a strategy would benefit significantly from the identification of putative host ligands that interact with vpr. Interestingly, recent work has identified several candidate ligands from biochemical evidence (24, 25). Among these, an approximately 41-kDa rip-1 protein that exhibited cytoplasmic and nuclear forms appears relevant to these discussion (24). More recently, through molecular technology, several additional putative ligands have been identified (41, 42) and are under investigation including uracil DNA glycosylase, of unclear association, and HHR23A, a cellular protein implicated in nucleotide excision DNA repair, a process important for cell growth.
In this article, we report the identification and molecular characterization of a human protein that specifically binds to Vpr and then colocates with vpr in the perinuclear region of the cell (Fig. 3). This cellular protein, which is identical to the concurrently identified human 34-kDa mov34 homologue (31), which we term hVIP/MOV34, displays a protein binding specificity predicted for an authentic cellular cofactor for HIV-1 Vpr. This profile includes a close correlation between the biological activity and hVIP/MOV34 interactions with a set of Vpr missense mutants (Fig. 4). We further show that hVIP/MOV34 Vpr interaction requires the carboxyl domain of Vpr for manifestation of its activity. Importantly, we observe that expression of hVIP/MOV34 antisense in mammalian cells can specifically arrest the cells at G2/M phase of the cell cycle (Fig. 5), supporting the hypothesis that hVIP/MOV34 is a necessary component for cell replication. A link in this case is that the loss of hVIP/MOV34 is associated with the inhibition of MPF-associated H1 kinase activity, which is essential for cells to enter into mitosis (Fig. 6). Although the data presented in this article argue for a role for hVIP/MOV34 in mediating some functions of HIV-1 Vpr, they shed light on its normal cellular function as well. The expression of hVIP/MOV34 in a wide variety of human tissues and diverse cell lines is strongly suggestive of an important biological role in the eukaryotic system.
The MOV34 gene was originally identified in mice after retroviral insertional mutagenesis induced by a recombinant Moloney leukemia provirus inserted into the mouse germ line (35). Embryos homologous for this proviral integration develop normally through the blastocyst stage but die shortly after implantation into the uterus. The product of the MOV34 gene, therefore, is an essential protein for embryonic viability. The human homologue (MOV34) maps to human chromosome 16q24 and is a member of a family of proteins that regulate transcription and the state of the host cell (32). This description is similar to the original definition of the effects of Vpr on host cell regulation of both transcription and proliferation, i.e., the differentiation state of the host cell (13). Important members of the MOV34 gene family include components of the eIF3 complex. eIF3 is a large multimeric complex that regulates transcriptional events and is essential for G1/S and G2/M phase progression through the cell cycle (32). The demonstration here that hVIP/MOV34 is important for cell cycle progression is consistent with such known functions of MOV family members and is similarly consistent with publications describing transcriptional effects of Vpr and its effects on cell proliferation (12–15). Furthermore, the capture of hVIP/MOV34 by Vpr at the nuclear membrane likely interferes with hVIP/MOV34 interactions with other host proteins that coordinately could regulate some aspects of G2/M transition.
In support of a complex interaction with other proteins, hVIP/MOV34 does contain nine dipeptide leucine/isoleucine motifs and six leucine/isoleucine repeats (Fig. 1). In general, such leucine/isoleucine repeats are believed to mediate protein–protein interactions (33, 34). The significance of these sequence elements in hVIP/MOV34 supports the hypothesis that, in a similar vein to other MOV34 family members, hVIP/MOV34 interacts with other host cell proteins to mediate its normal cellular function and allow transversion through the cell cycle. Immunofluorescence analysis of the intracellular localization of hVIP/MOV34 demonstrates that hVIP/MOV34 is concentrated in the cell nucleus and redirected to the nuclear membrane in the presence of Vpr (Fig. 3), demonstrating that hVIP/MOV34 and Vpr interact in vivo. These observations do not allow us to make conclusions of the role of other host proteins in this process.
Thus, these observations demonstrate that Vpr specifically interacts with an important host protein, hVIP/MOV34, which is likely to be part of a larger protein complex that may be a component of the mammalian cell cycle cascade. The ability of Vpr to inhibit cellular proliferation is possibly mediated by redirection of hVIP/MOV34 from the nucleus to the nuclear periphery, possibly preventing interaction of hVIP/MOV34 with its natural ligand(s) and inhibiting MPF-associated kinase activity. The relationship of other putative ligands of Vpr and this ligand will require further exploration. The diverse expression pattern of hVIP/MOV34 supports that this protein has general relevance to cell division in multicellular organisms. Furthermore, these studies begin to elucidate a pathway through which HIV-1 Vpr may mediate specific control of cellular function, thus presenting opportunities for both development of HIV therapeutics and potential anticancer strategies.
Acknowledgments
We thank Dr. Jean Bennett for assistance with the immunofluorescence studies and Dr. Rick Ciccarelli for reagents important to these studies. Additionally we thank Suman, Rebecca, Lindsey, and Elizabeth for support and critical discussions. This work was supported in part by grants from the National Institutes of Health to D.B.W. and GM47439 to R.J.M.
ABBREVIATIONS
- hVIP
human Vpr interacting protein
- MPF
maturation promoting factor
- PBMC
peripheral blood mononuclear cell
- eIF3
eukaryotic initiation factor 3
References
- 1.Adachi A, Ono N, Sakai H, Ogawa K, Shibata R, Kiyomasu T, Masuike H, Ueda S. Arch Virol. 1991;117:45–58. doi: 10.1007/BF01310491. [DOI] [PubMed] [Google Scholar]
- 2.Akari H, Sakuragi J, Takebe Y, Tomonaga K, Kawamura M, Fukasawa M, Miura T, Shinjo T, Hayami M. Arch Virol. 1992;123:157–167. doi: 10.1007/BF01317146. [DOI] [PubMed] [Google Scholar]
- 3.Cohen E A, Terwilliger E F, Jalinoos Y, Prouix J, Sodroski J G, Haseltine W W A. J Acquired Immune Defic Syndr. 1990;3:11–18. [PubMed] [Google Scholar]
- 4.Dedera D, Hu W, Vander Heyden N, Ratner L. J Virol. 1989;63:3205–3208. doi: 10.1128/jvi.63.7.3205-3208.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa K, Shibata R, Kiyomasu T, Kiguchi I, Kishida Y, Ishimoto A, Adachi A. J Virol. 1989;63:4110–4114. doi: 10.1128/jvi.63.9.4110-4114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrigo S J, Chen I S Y. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 7.Garrett E D, Tiley L S, Cullen B R. J Virol. 1991;65:1653–1657. doi: 10.1128/jvi.65.3.1653-1657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen B R. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian R A, Cohen E A. J Virol. 1994;68:6831–6835. doi: 10.1128/jvi.68.11.6831-6835.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinzinger N K, Bukrinsky M I, Haggery S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy D N, Fernandes L S, Williams W V, Weiner D B. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-y. [DOI] [PubMed] [Google Scholar]
- 12.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams W V, Green D R, Weiner D B. Nat Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 13.Levy D N, Rafaeli Y, Weiner D B. J Virol. 1995;69:1243–1252. doi: 10.1128/jvi.69.2.1243-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingam S, MacDonald B, Ugan K E, Ayyavoo V, Agadjayan M G, Williams W V, Weiner D B. DNA Cell Biol. 1997;16:137–143. doi: 10.1089/dna.1997.16.137. [DOI] [PubMed] [Google Scholar]
- 15.Rogel M E, Wu L I, Emerman M. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Re F, Braaten D, Franke E K, Luban J. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mustafa F, Robinson H. J Virol. 1993;67:6909–6915. doi: 10.1128/jvi.67.11.6909-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy D N, Refaeli Y, Weiner D B. Curr Top Microbiol Immunol. 1995;193:209–236. doi: 10.1007/978-3-642-78929-8_11. [DOI] [PubMed] [Google Scholar]
- 20.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 22.Balotta C, Lusso P, Crowley R, Gallo R C, Franchini G. J Virol. 1993;67:4409–4414. doi: 10.1128/jvi.67.7.4409-4414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 24.Refaeli Y, Levy D N, Weiner D B. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L J, Mukherjee S, Narayan O. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 26.Altschul S F, Warren G, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Fuerst T R, Earl P L, Moss B. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahalingam S, Collman R G, Patel M P, Monken M P, Srinivasan A. Virology. 1995;212:331–339. doi: 10.1006/viro.1995.1490. [DOI] [PubMed] [Google Scholar]
- 29.Pines J, Hunter T. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields S, Song S A. Nature (London) 1989;340:245–247. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 31.Asano K, Vornlocher H, Richter-Cook N J, Merrick W C, Hinnebusch A G, Hershey J W B. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 32.Verlhac M H, Chen R-H, Hanachi P, Hershey J W B, Derynck R. EMBO J. 1997;16:6812–6822. doi: 10.1093/emboj/16.22.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki N. Proc Natl Acad Sci USA. 1990;92:3794–3798. [Google Scholar]
- 34.Draper M P. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gridley T, Jaenisch R, Gendron-Maguire M. Genomics. 1991;11:501–507. doi: 10.1016/0888-7543(91)90056-k. [DOI] [PubMed] [Google Scholar]
- 36.Gerhart J, Wu M, Krischner M W. J Cell Biol. 1994;98:1247–1255. doi: 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boocher R N, Alfa C E, Hyams J S, Beach D H. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 38.Gautier J, Norbury C, Lohka M, Nurse P, Maller J. Cell. 1988;54:433–439. doi: 10.1016/0092-8674(88)90206-1. [DOI] [PubMed] [Google Scholar]
- 39.Miake-Lye R, Newport J, Krischner M W. J Cell Biol. 1983;97:81–91. doi: 10.1083/jcb.97.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 41.BouHamdan M, Benichou S, Rey F, Navarro J M, Agostini I, Spire B, Camonis J, Sluppaug G, Vigne R, Benarous R, Sire J. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poon B, Jowett J B, Stewart S A, Armstrong R W, Rishton G M, Chen I S. J Virol. 1997;71:3961–3971. doi: 10.1128/jvi.71.5.3961-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]