Abstract
Cryptococcus neoformans is a fungal pathogen that, after inhalation, can disseminate to the brain. Host alveolar macrophages (AMs) represent the first defense against the fungus. Once phagocytosed by AMs, fungal cells are killed by a concerted mechanism, involving the host-cellular response. If the cellular response is impaired, phagocytosis of the fungus may be detrimental for the host, since C. neoformans can grow within macrophages. Here, we identified a novel cryptococcal gene encoding antiphagocytic protein 1 (App1). App1 is a cryptococcal cytoplasmic protein that is secreted extracellularly and found in the serum of infected patients. App1 does not affect melanin production, capsule formation, or growth of C. neoformans. Treatment with recombinant App1 inhibited phagocytosis of fungal cells through a complement-mediated mechanism, and Δapp1 mutant is readily phagocytosed by AMs. Interestingly, the Δapp1 mutant strain showed a decreased virulence in mice deficient for complement C5 (A/Jcr), but it was hypervirulent in mice deficient for T and NK cells (Tgε26). This study identifies App1 as a novel regulator of virulence for C. neoformans, and it highlights that internalization of fungal cells by AMs increases the dissemination of C. neoformans when the host cellular response is impaired.
Introduction
Cryptococcosis is a chronic human disease caused by the fungus Cryptococcus neoformans. The disease occurs after inhalation of encapsulated yeasts or basidiospores into the alveolar spaces and eventually develops with the dissemination of C. neoformans to the central nervous system, since the fungus is neurotropic, causing the most common fungal meningoencephalitis worldwide (1). The majority of cryptococcosis cases have been reported in individuals with impaired immunity. The exact immune defect(s) that favor cryptococcosis are not known, but evidence suggests that an intact complement system (2), macrophage defense mechanisms (3), and/or specific cell-mediated immunity (T and NK cells) are necessary to prevent the dissemination of the infection (4–6).
On the other hand, C. neoformans may produce virulence factors to evade host defenses and cause progressive disease. Several factors have been associated with virulence, including capsule production, melanin formation, and the ability to grow at 37°C. Various signaling pathways regulate these virulence factors, such as those mediated by calcineurin, G proteins (1), and, as demonstrated more recently, by the phospholipid and sphingolipid pathways (7–9). In particular, in our previous studies we demonstrated that modulation of C. neoformans inositol phosphoryl ceramide synthase 1 (Ipc1), an essential enzyme of the sphingolipid pathway, regulates not only melanin formation but also the ability of fungal cells to grow, once ingested, inside macrophages (9).
Cryptococcus neoformans is a facultative intracellular pathogen (10, 11). The ability of C. neoformans to produce disease is therefore a combination of both intracellular and extracellular growth in the infected host. Five different stages have been suggested for C. neoformans infection: initiation, dormancy, reactivation, dissemination, and proliferation (1). Since the lung is the port of entry for C. neoformans, recruited alveolar macrophages (AMs) represent one of the cell types that inhibit and kill C. neoformans (12, 13). During the initiation process, T cells are activated, and through the release of macrophage-activating cytokines they promote the formation of granulomas, resulting in the destruction of the intracellular fungus or containment in a latent state (dormancy) (11). In individuals with impaired cellular immunity, C. neoformans grows intracellularly, eventually lysing the macrophage (reactivation-dissemination). The released organism can then infect other phagocytes, increasing the intracellular growth of the fungus (dissemination-proliferation). On the other hand, C. neoformans can also grow extracellularly, eluding phagocytosis through the production of specific fungal factor(s) that disable the recognition and response of the host immune system. Which component of the cryptococcal population, intracellular or extracellular, most affects the progression of the disease is still unclear (14–18).
Here, we have identified a novel virulence factor of C. neoformans regulated by Ipc1 that specifically inhibits the phagocytosis of yeast cells by AMs. Because of its distinctive function, we named this fungal factor antiphagocytic protein 1 (App1). We characterized the Ipc1-App1 pathway in the regulation of phagocytosis of C. neoformans by AMs using a combination of genetic and pharmacological approaches. We found that App1 inhibits attachment and ingestion of yeast cells by AMs through a complement-mediated mechanism. Interestingly, by testing Ipc1 and App1 mutant strains in C5 and T-NK immunodeficient mouse models, we found that whereas Ipc1 is required for the dissemination of C. neoformans from the lung to the brain in both animal models, lack of App1 favors the dissemination of yeast cells within macrophages when the T and NK cellular immunity is altered.
Methods
Strain and growing media.
The following strains were used in this study: the Cryptococcus neoformans variety grubii serotype A strain H99 (WT), the M001 strain, an ade2 isogenic derivative of H99, GAL7:IPC1 strains 10 and 13 (created from M001) (9), two independent Δapp1 knockout strains (6 and 31, also created from M001), the IPC1Rec strain (created from GAL7:IPC1 10), and the Δapp1Rec strain (created from Δapp1 31). A synthetic medium, containing 6.7 g/l yeast nitrogen base (YNB) without amino acids, 1.3 g/l amino acid mix lacking adenine, 180 g/l sorbitol, 20 g/l glucose, and 20 g/l agar, was used for selecting the Δapp1 6 and 31 strains obtained from biolistic transformation. C. neoformans H99, M001, GAL7:IPC1, and Δapp1 strains were routinely grown on yeast peptone dextrose (YPD) medium. Yeast extract peptone (YP) supplemented with 20 g/l glucose or 20 g/l galactose was used for down- and upregulation of IPC1 expression, respectively. YP agar plates supplemented with 2% glucose and 200 U/ml hygromycin B (Calbiochem, San Diego, California, USA) were used to select and screen both IPC1Rec and Δapp1Rec strains. Antibody-coated erythrocytes (IgM/IgG) are from Advance Research Technologies (San Diego, California, USA). Complement/antibody-coated erythrocytes were created by incubating the IgM/IgG-coated erythrocytes with C5-deficient human serum (a gift from Stephen Tomlinson, Medical University of South Carolina, Charleston, South Carolina, USA) for 1 hour at 37°C. Sera of patients with AIDS and normal subjects were a gift from Angela Restrepo (Medellin, Colombia).
Collection and preparation of mouse AMs.
A/Jcr (NCI/Frederick Laboratories, Frederick, Maryland, USA) and transgenic epsilon 26 (Tgε26) mice (19) were used to obtain AMs. Animals were euthanized by CO2 inhalation. This procedure does not affect the phagocytic ability of AMs (20). Then, the trachea was exposed, and bronchoalveolar lavage (BAL) was performed by inserting a 20-gauge Luer stub adapter attached to a 1-ml syringe into the trachea. One milliliter of warm HBSS with 50 U/ml penicillin-streptomycin was injected into the lung and recovered. The BAL was repeated 10 times, and recovered fluid was pooled for assessment of cellular content. From each mouse, a total of 3–5 × 105 AMs were obtained. Total cell count was assessed in a standard hemocytometer using a cell suspension diluted 1:1 with trypan blue. A total of 3 × 104 AMs were plated into each well of a 96-well microtiter plate (Becton Dickinson, Franklin Lakes, New Jersey, USA) in 100 μl of DMEM (Invitrogen Life Technology, Carlsbad,California, USA) containing either glucose or galactose with 10% fetal calf serum, 1% L-glutamine, and 100 U/ml penicillin-streptomycin and incubated at 37°C (5% CO2) for 2 hours. Nonadherent AMs were removed by washing the wells with 100 μl of warm medium. When the bronchoalveolar lavage was completed, a 22-gauge needle attached to a 1-ml syringe was inserted into the heart, and blood was collected. Blood was then put in Vacutainer blood collection tubes containing no additives (Becton Dickinson Vacutainer System) to obtain normal serum. After centrifugation, serum was collected and used to opsonize yeast cells.
Phagocytosis assay.
C. neoformans WT H99 and GAL7:IPC1 (10 and 13) strains were grown in YPD for 16 hours at 30°C in a shaker incubator at 250 rpm. Cells were washed twice with sterile PBS (pH 7.4), resuspended, and diluted into 10 ml of fresh YP broth with either glucose or galactose to a final density of 105 cells per milliliter and incubated for 24 hours at 30°C in a shaker incubator at 250 rpm. C. neoformans Δapp1 mutant 6 and 31 strains were grown in YP with glucose for 24 hours at 30°C in a shaker incubator at 250 rpm. Cells were washed twice with PBS and resuspended in DMEM (glucose or galactose) containing 10% fresh mouse serum. Next, 6 × 105 opsonized yeast cells were added to AMs, and plates were incubated at 37°C (5% CO2) for 2 hours. IgM/IgG-coated or iC3b-IgM/IgG–coated erythrocytes were diluted in DMEM, and 6 × 106 cells were added to AMs. Nonattached yeast or erythrocyte cells were then removed by washing with PBS. Plates were placed under an inverted microscope, and the phagocytic index was defined as the percentage of yeast cells attached or ingested per number of macrophages per field, as previously described (9). For each experiment, at least four fields were observed in three different wells per strain or treatment condition. Results are expressed as geometric means ± SDs of the phagocytic indexes obtained for each experiment. Statistical analysis was performed with Student’s t test. A P value of less than 0.05 was considered to be significant. For ex vivo phagocytosis, mice were anesthetized with an intraperitoneal injection of 60 μl of xylazine/ketamine mixture, containing 95 mg of ketamine per kilogram of body weight and 5 mg of xylazine per kilogram of body weight. Then, C. neoformans WT, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains were inoculated intranasally. Three mice were used for each yeast strain. After 2 hours, mice were euthanized by CO2 inhalation, and AMs and yeast cells were collected by BAL. Cells were then centrifuged at 1,200 rpm and resuspended in 500 μl of PBS, and 10 μl was plated in a standard hemocytometer for the determination of the number of yeast cells attached and/or ingested by AMs (phagocytic index). Results represent the geometric means ± SDs of the phagocytic indexes obtained from three animals per strain.
Differential display RT-PCR.
Differential display was performed according to the instructions for the GenHunter RNA-Image Kit (GenHunter Corp., Nashville, Tennessee, USA). Total RNA, obtained from C. neoformans WT H99 and GAL7:IPC1 strain 10 grown on glucose or galactose, was divided into three equal aliquots, and first-strand synthesis was performed using oligo-dT anchored with an A, C, or G. This cDNA was then subjected to PCR amplification with an arbitrary primer (AP) and the corresponding oligo-dT-A, -dT-C, or -dT-G, respectively. Four arbitrary primers were used (AP1, AP2, AP3, and AP4) for each oligo-dT, producing 12 different amplifications of gene transcription patterns for each strain (WT-glucose, GAL7:IPC1-glucose, WT-galactose, and GAL7:IPC1-galactose). For radioactive labeling, α-[33P]dATP was added in each PCR reaction. PCR products were separated at 1000 V in a 6% polyacrylamide gel. The gel was then transferred onto a 3M paper, dried, marked in each corner with radioactive ink, and exposed to Kodak X-Omat film. Bands of interest were identified as those that were overexpressed in GAL7:IPC1-galactose as compared with WT-galactose and downregulated in GAL7:IPC1-glucose as compared with WT-glucose, with no significant difference in expression between WT-glucose and WT-galactose. Once the bands had been identified, the film was oriented to the dried gel, and the corresponding slide was cut out with a razor blade. Gel slides were eluted, and DNA fragment was precipitated with 100% ethanol and reamplified with PCR using the corresponding primers. PCR product was then separated in a regular 1% agarose gel stained with ethidium bromide, and fragment(s) of interest were extracted and cloned into the PCR-TRAP Cloning System (GenHunter Corp.). The plasmid was then amplified, reconfirmed by PCR to carry the fragment of interest, and sequenced.
Ipc1 activity, RT-PCR, and Western blot.
Ipc1 activity was performed as previously described (9). RT-PCR directed against the APP1 gene was performed using 1 μg of total RNA for each group (WT-glucose, GAL7:IPC1-glucose, WT-galactose, and GAL7:IPC1-galactose) following the protocol described by the SUPERSCRIPT First-Strand Synthesis System for RT-PCR (Invitrogen, Life Technology). First-strand synthesis of APP1 gene was made with primer IP13 (5′-AAT CAT CAA TGT TCG CAG CTC CTTC-3′). Second-strand synthesis of APP1 gene was with primers IP13 and IP15 (5′-ATG ATG TCC TCT GCC ACT GCT GAAC-3′), yielding a 543-bp fragment. Actin-specific primers AC-2 (5′-CAG CTG GAA GGT AGA CAA AGA GGC-3′) and AC-1 (5′-CGC TAT CCT CCG TAT CGA TCT TGC-3′) were used for the actin gene as a control, yielding a 543-bp fragment.
For Western blot, proteins from 24-hour cultures of WT-glucose, GAL7:IPC1-glucose, WT-galactose, and GAL7:IPC1-galactose were extracted as previously described (9), electrophoresed in 10% SDS-PAGE, and transferred onto a nitrocellulose membrane. Membrane was then blocked with 5% nonfat milk/1× PBS/0.1% Tween 20 (5% NFM/PBST). The primary antibody was anti-App1 at a 1:1,000 dilution in 5%NFM/PBST. The rabbit secondary antibody was anti-IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, California, USA) in 5%NFM/PBST. Membrane was washed generously in 5%NFM/PBST, treated with ECL (Amersham Pharmacia Biotechnology, Piscataway,New Jersey, USA), and exposed to Kodak X-Omat film. Analysis of Ipc1 activity and APP1 mRNA and protein levels was performed in two independent GAL7:IPC1 transformants (nos. 10 and 13; see Figure 2).
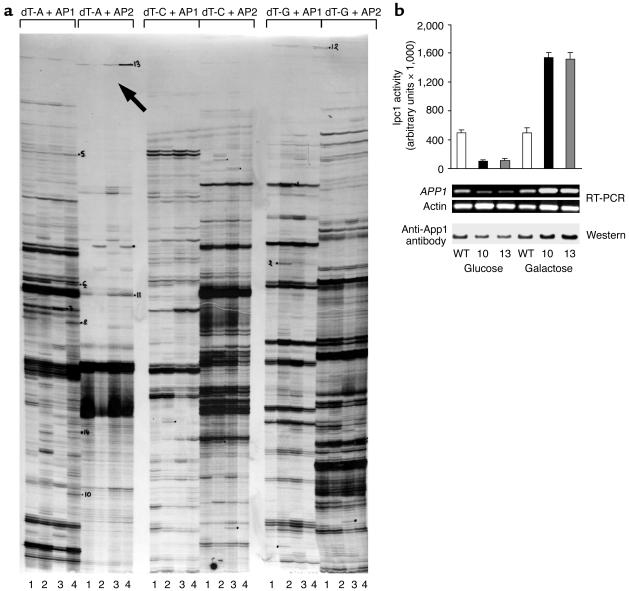
Figure 2.
(a) DD-RT-PCR in WT and GAL7:IPC1 10 strains using dT-A, -C, and -G with AP1 and AP2 primers. The number 1 denotes WT-glucose; 2, GAL7:IPC1-glucose; 3, WT-galactose; and 4, GAL7:IPC1-galactose. The black arrow indicates the App1 band, which is downregulated when Ipc1 is downregulated (no. 2) and upregulated when Ipc1 is upregulated (no. 4), with no significant changes in WT-glucose (no. 1) or WT-galactose (no. 3). (b) Ipc1 activity, RT-PCR, and Western blot of App1 in C. neoformans WT H99 and GAL7:IPC1 strains grown on glucose and galactose medium. APP1 expression is significantly regulated by Ipc1 modulation. Actin gene was used as a control. Two independent GAL7:IPC1 strains were used for RT-PCR (nos. 10 and 13).
Disruption of the APP1 gene.
A disruption construct was made (see Figure 3) by using a series of molecular manipulation involving the APP1 gene and the 3.0-kb ADE2 marker genomic DNA fragment obtained from serotype A, strain H99. First, fragment A (784 bp) was generated by PCR using H99 genomic DNA as a template and primers IP(XhoI) (5′-GTTCTCGAGCTTTGGGAGATGGCGGCTCACTTTG-3′) and IP(Eco-1) (5′-CTATGAATTCGTATTATTTGTTGGGTTGGCTTAC-3′), containing a XhoI and EcoRI site, respectively (bolded and underlined). The 784-bp fragment contains sequence corresponding to the 5′ upstream untranslated region (5′-UTR) of the APP1 open reading frame (ORF) (see Figure 3). This fragment was then digested with XhoI and EcoRI, producing fragment B (775 bp). Second, fragment C (888 bp) was generated using H99 genomic DNA as a template and primers IP(XbaI) (5′-CAATCTAGACATGCGTATAGTACGCCGGTTGAAG-3′) and IP(Eco-2) (5′-CAATGAATTCGTATATCTCTGTACTATTC-3′), containing an XbaI and EcoRI site, respectively (bolded and underlined). This fragment was then digested with XbaI and EcoRI, producing fragment D (879 bp), which contains sequence homologous to the 3′ downstream untranslated region (3′-UTR) of APP1 ORF (see Figure 3). Third, fragment B (5′-UTR) was combined with fragment D (3′-UTR), ligated, and cloned into the XhoI and XbaI sites of the SK+ pBluescript vector, generating plasmid pΔapp1. The pΔapp1 plasmid was sequenced to make sure that no mutations were introduced by PCR manipulations. Finally the EcoRI-restricted ADE2 fragment was inserted into the EcoRI site located between 5′-UTR and 3′-UTR fragments to form the pΔapp1/ADE2 disruption construct. The pΔapp1/ADE2 was transformed into C. neoformans strain M001, using biolistic delivery of DNA, following the protocol as described by Toffaletti et al. (21). Transformants were grown on YNB-glucose without adenine (see above). Forty stable transformants were chosen, and genomic DNA was extracted and assayed for PCR and/or Southern blot analysis according to Sambrook et al. (22). Transformants showing integration of the pΔapp1/ADE2 construct at the APP1 locus were chosen and designed Δapp1 strains.
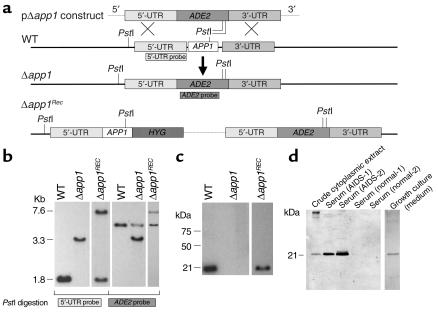
Figure 3.
Disruption of APP1 gene. (a) Schematic representation for disruption and reconstitution of APP1 gene. (b) Southern analysis. Genomic DNA from C. neoformans WT H99, Δapp1, and Δapp1Rec strains were extracted, digested with PstI, electrophoresed in a 0.7% agarose gel, transferred to a Nytran membrane, and blotted against 5′-UTR and ADE2 probes. The Δapp1 strain showed a deletion of the APP1 gene with insertion of the ADE2 gene. Reintroduction of the APP1 gene in the APP1 locus is shown in the Δapp1Rec strain. (c) Western analysis of cell lysate from WT, Δapp1, and Δapp1Rec cell cultures using rabbit anti-App1 polyclonal antibodies. App1 protein is absent in the Δapp1 mutant and reconstituted in the Δapp1Rec strain. (d) Western analysis of crude cytoplasmic extract, serum of two AIDSpatients, serum from two normal subjects, and growth culture medium. App1 is secreted extracellularly in vitro and during cryptococcal meningoencephalitis. HYG, hygromycin B gene.
Reconstitution of the IPC1 and APP1 genes in the GAL7:IPC1 and Δapp1 mutant strains.
The IPC1 gene expression in the GAL7:IPC1 strains is under the control of the GAL7 promoter, and its activity is modulated by glucose or galactose. To reconstitute the expression of IPC1, the GAL7 promoter was replaced with the IPC1 promoter using a plasmid cassette containing the upstream UTR of the IPC1 locus, the ADE2 gene, the hygromycin B gene, the IPC1 promoter, and the 5′-truncated IPC1 gene. This plasmid cassette was introduced into the GAL7:IPC1 strain 10. Hygromycin B–resistant transformants were selected and screened for homologous recombination at the IPC1 locus. Transformant 5 was selected and showed that Ipc1 activity is no longer modulated by glucose or galactose and is similar to the WT strain (data not shown). This transformant was named the IPC1Rec strain. To reconstitute the Δapp1 mutant strain, the WT APP1 gene was reintroduced into the Δapp1 mutant 31 strain by transforming the APP1 promoter fused to the APP1 gene linked to a HYGB gene conferring hygromycin B resistance (23). Genomic DNA was isolated from six hygromycin B–resistant transformants and analyzed by Southern blot. Transformant 4 showed that both the Δapp1:ADE2 mutant allele and the WT APP1 gene were now present in the APP1 locus (see Figure 3) and that App1 protein was now produced (see Figure 3). This transformant was named the Δapp1Rec strain.
Production of App1 recombinant protein.
App1 recombinant proteins were produced from C. neoformans serotype A using the corresponding APP1 cDNA. The pAPP1-cDNA plasmid from serotype A strain H99 was used as template for PCR with primers Dan5 (5′-CATCTCGAGTGCCACTGCTGAACTT-3′) and Dan3 (5′-CATGAATTCTCAAATCATCAATGT-3′), containing XhoI and EcoRI sites, respectively (bolded and underlined). The resulting fragment was then digested with XhoI and EcoRI and cloned into XhoI-EcoRI–restricted pBAD-HIS-B vector (Invitrogen, Life Technology), generating pBAD-His-App1. This vector contains App1 cDNA in frame with Xpress and HIS tags at the 5′ region of App1 gene. The pBAD-His-App1 vector was sequenced to ensure that no mutations were inserted by genetic manipulations and then transformed into E. coli TOP10 strain (Invitrogen, Life Technology). Induction of App1 proteins was obtained following the protocol described by the Invitrogen expression system. After an overnight culture on Luria Bertani (LB) medium containing 100 μg/ml ampicillin, induction of App1 was obtained by growing E. coli cells for an additional 4 hours at 37°C in 80 ml LB medium containing 50 μg/ml ampicillin and 2% arabinose in a shaker incubator at 250 rpm. Cells were then collected by centrifugation at 6,000 rpm and resuspended in lysis buffer (20 mM NaPO4 [pH 7.4], 20% glycerol, 1 mM PMSF, and 1 mg/ml lysozyme). After 30 minutes of incubation on ice, cells were lysed by sonication (six cycles of 1 minute each, 20% power). Cell lysate was then centrifuged at 20,000 g for 30 minutes at 4°C, and supernatant was further diluted with 20 mM sodium phosphate (pH 7.8, buffer A) and applied to a HiTrap chelating HP column (Amersham) (0.2 ml per minute) previously charged with 0.1 M NiSO4 according to the manufacturer’s instructions and equilibrated with buffer A. The column was then washed with five column volumes of buffer A, and bound proteins were eluted at 1 ml per minute with an increasing linear gradient of 20 mM sodium phosphate containing 0.5 M imidazole (pH 7.8, buffer B). Fractions were analyzed by Western blotting, and those with the highest amount of App1 (usually at 0.1–0.2 M imidazole) were combined (fractions 36–42), the buffer exchanged with 20 mM Tris-HCl (pH 8), 1 mM EDTA, 0.2 mM PMSF, and 0.005% Triton X-100 (buffer C) using PD-10 columns (Amersham) and applied to a HiTrap Q previously equilibrated with buffer C. After the column was washed with five column volumes of buffer C, bound proteins were eluted with an increasing linear gradient of buffer C containing 1.5 M NaCl, and fraction Q54 — with the highest ratio of App1 to contaminating proteins — was selected and used for cellular treatments. The lysate from cells expressing pBAD-His (mock) vector was used as a control and subjected to a purification protocol identical to the one used with pBAD-His-App1.
Virulence studies in murine models of cryptococcal meningitis.
Four- to six-week-old male Tgε26 mice (Edward Balish, Medical University of South Carolina, Charleston, South Carolina, USA), and four- to six-week-old female A/Jcr mice (NCI/Frederick Laboratories) were used. The Tgε26 mice lack functional T and NK cells, whereas macrophages, monocytes, and granulocytes are present and functional (19, 24). The A/Jcr model from NCI/Frederick Laboratories has a slight C5 deficiency (25), and, as a consequence, they mount a predominant Th2 response (26–28). Mice were anesthetized with an intraperitoneal injection of 60 μl of xylazine/ketamine mixture, containing 95 mg of ketamine per kilogram of body weight and 5 mg of xylazine per kilogram of body weight. Wild-type H99 and the isogenic GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains were prepared by growing yeast cells for 24 hours at 30°C in YPD medium. The cells were pelleted, washed twice, and resuspended in PBS (pH 7.4) at a concentration of 2.5 × 106 cells per milliliter. Ten A/Jcr and 14 Tgε26 mice were infected with 5 × 104 cells of the H99, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains in a volume of 20 μl through nasal inhalation. The mice were fed ad libitum and followed with twice-daily inspections. Mice that appeared moribund or in pain were sacrificed using CO2 inhalation. Survival data from the mice experiment were analyzed using the Kruskal-Wallis test. A P value of less than 0.05 was considered to be significant.
Tissue-burden culture studies in murine models of cryptococcosis.
Fifteen mice of each model were infected with 5 × 104 cells each of WT, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains in a volume of 20 μl through nasal inhalation. C. neoformans strains were prepared as for the virulence studies. The mice were fed ad libitum and followed with twice-daily inspections. At selected time points after inoculation, five mice for each strain were sacrificed with CO2 inhalation, and lungs and brains were removed, weighed, and homogenized in 10 ml sterile PBS using Stomacher 80 (Lab System, Fisher Scientific, Pittsburgh, Pennsylvania, USA) for 120 seconds at high speed. Serial dilutions were then plated onto YPD plates in triplicate. Plates were incubated at 30°C for 72 hours, yeast colonies were counted, and the number was recorded as the geometric mean(s) of the logarithm of CFU per organ. Statistical analysis was performed using Student’s t test, and a P value of less than 0.05 was considered to be significant. We also must note that two independent mutants for Ipc1 (GAL7:IPC1 10 and 13) and App1 (Δapp1 6 and 31) were used for both survival and tissue-burden cultures studies. No significant differences were found between GAL7:IPC1 10 and 13 strains and between Δapp1 31 and 6 strains (data not shown).
Histology studies in murine models of cryptococcosis.
Two mice of each model were infected with 5 × 104 cells each of WT, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains in a volume of 20 μl through nasal inhalation. C. neoformans strains were prepared as for the virulence and tissue-burden studies. At selected time points (day 5 and 15 after infection for A/Jcr mice and day 6 and 13 after infection for Tgε26 mice), lungs and brains were removed, placed in 10% formalin, and then embedded in paraffin. Sections were stained with periodic acid–Schiff (PAS) to identify fungal cell wall, with mucicarmine as an additional staining for the identification of C. neoformans capsule (data not shown), with hematoxylin and eosin to examine the host inflammatory response, and with anti-CD68 antibody to identify macrophages. Sections were examined by light microscopy.
Results
Ipc1 regulates phagocytosis of C. neoformans by alveolar macrophages.
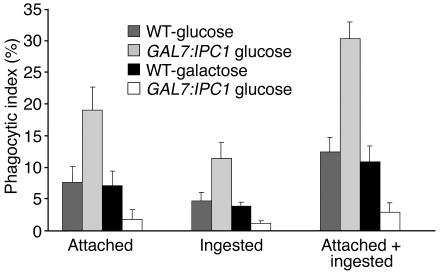
In our previous studies, we found that once C. neoformans is internalized by the macrophage-like cell J774.16, its intracellular growth is regulated by Ipc1 expression (9). Here, we investigated whether the level of Ipc1 expression would affect the phagocytosis of C. neoformans by mouse AMs. Phagocytosis (attached, ingested, and attached plus ingested) was increased by 2.4-fold when Ipc1 was downregulated (GAL7:IPC1 grown on glucose, P = 0.005) and decreased by 2.5-fold when Ipc1 was upregulated (GAL7:IPC1 grown on galactose, P = 0.004). Similar results were obtained with a second and independent GAL7:IPC1 transformant 13 (data not shown). No difference was observed between WT cells grown on glucose or galactose (Figure 1). Additionally, by testing the IPC1Rec strain, we found no significant difference in attached and/or ingested yeast cells with the WT strain (data not shown).
Figure 1.
Phagocytosis assay of C. neoformans WT H99 and GAL7:IPC1 10 strains grown on glucose or galactose medium. Decrease of Ipc1 increases phagocytosis, whereas increase of Ipc1 decreases phagocytosis (P = 0.004 and 0.003, respectively). Phagocytic index is expressed as the geometric mean of the percentage of yeast cells attached or ingested per number of macrophages per field.
Ipc1 regulates the expression of App1.
In order to determine the mechanism(s) by which Ipc1 regulates phagocytosis, we sought for downstream effector(s) of Ipc1 by differential display RT-PCR (DD-RT-PCR). C. neoformans WT H99 and GAL7:IPC1 10 strains were grown under Ipc1-repressing (glucose) or Ipc1-inducing (galactose) conditions, and differentially expressed bands were identified in a polyacrylamide gel. Four bands that were up- and downregulated when Ipc1 was up- and downregulated, respectively, as compared with the corresponding WT grown on glucose or galactose, were selected. Three bands showed no homology when their sequences were blasted in the GenBank database (data not shown). Interestingly, one fragment (Figure 2a, black arrow) produced 100% homology with inhibitory protein 1 (IP1) (GenBank accession numbers AF180107, AF180108, and AY101600). This protein and the corresponding gene were previously isolated from C. neoformans by our group using different methods (unpublished observations). IP1 is a cytoplasmic factor that is secreted into the growth medium, and it inhibits phagocytosis of C. neoformans by AMs (29). We renamed the previously isolated IP1 antiphagocytic protein 1 (App1). The finding that Ipc1 modulation regulates the gene expression of APP1 was then validated by RT-PCR using specific primers against the APP1 gene and by Western analysis using anti-App1 polyclonal antibodies. Indeed, App1 was significantly up- and downregulated when Ipc1 expression was modulated as compared with the WT cells (Figure 2b).
APP1 is a unique gene of C. neoformans.
The sequence analysis obtained from the H99 genomic and cDNA (GenBank accession number AF180108) revealed the presence of two intronic sequences of 78 and 63 bp, producing an ORF of 543 nucleotides encoding for 181 amino acids. A putative TATA box (TATAAAA) was identified at position –72 and a possible CAT box (CAAAT) at position –10 from the ATG start site. A potential polyadenylation site (ATAAAA) was identified at position +177 from the stop codon. Sequence analysis of APP1 revealed no significant homology with other DNA or proteins in the GenBank databases. Analysis of App1 amino acid sequences revealed the presence of five recognition sequence motifs, identified by Prosite Patterns (30). These predicted motifs comprised five putative phosphorylation sites (four for protein kinase C and one for tyrosine kinase), three myristoylation sites, and one glycosylation site. It appears, however, that a posttranslational modification is not required for App1 activity, since recombinant App1 protein produced using a prokaryotic expression system is active (see below). Whether a posttranslational modification(s) may modulate App1 activity awaits further studies. No other sequence information was obtained from homology searches.
Disruption of the APP1 gene and its role in phagocytosis.
To determine the role of the APP1 gene in the regulation of phagocytosis of C. neoformans, we disrupted the APP1 gene using the pΔapp1/ADE2 plasmid cassette. Out of forty ADE+ transformants, two transformants showed that the APP1 ORF had been replaced by the ADE2 gene. Transformant 31 showed a double crossover event (Figure 3b). The homologous recombination was then confirmed by PCR using two primers flanking the upstream and downstream sequence of APP1 ORF (data not shown). Transformant 31 was chosen and named the Δapp1 mutant strain (Figure 3). Subsequently, the absence of APP1 mRNA and App1 protein in the Δapp1 mutant strain was confirmed by Northern (data not shown) and Western (Figure 3c) blotting. Reconstitution of the Δapp1 mutant was performed by the reintroduction of the APP1 gene into the Δapp1 31 strain, and transformants were analyzed by Southern and Western blotting. As shown in Figure 3b, the APP1 gene was reintroduced in the APP1 locus, and production of App1 protein was restored (Figure 3c). The characterization of the Δapp1 strain revealed that the absence of App1 did not affect capsule formation, melanin production, and growth at 30°C or 37°C at either pH 7.0 or 4.0 (data not shown).
Interestingly, we found that App1 protein is secreted extracellularly, in the growth medium of C. neoformans, demonstrating that App1 is a secreted protein (Figure 3d). App1 secretion was absent in the Δapp1 mutant and was restored in the Δapp1Rec strain (data not shown). Since App1 was found in the growth medium of C. neoformans cultures, we wondered whether App1 would be secreted in vivo during the infection by C. neoformans. We therefore tested the serum of two patients with AIDS and cryptococcosis and the serum of two normal subjects for the presence of App1. As shown in Figure 3d, App1 was detected in sera of two patients with AIDS affected by cryptococcal meningoencephalitis, whereas it was absent in sera of normal subjects. Although from this experiment it is not known when App1 is secreted most during the infection by C. neoformans (e.g. initiation, dormancy, reactivation, dissemination, or proliferation), our data clearly show that App1 is produced and secreted (Figure 3d). The App1 secretion in vivo may be important for the development of cryptococcosis by affecting the phagocytosis of C. neoformans.
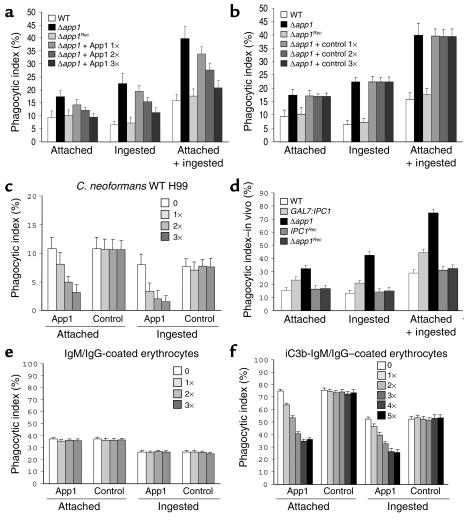
Therefore, we tested whether phagocytosis of C. neoformans was affected by the absence of App1. C. neoformans WT H99, Δapp1, and Δapp1Rec strains were incubated with AMs, and the phagocytic indexes were determined. As shown in Figure 4a, the lack of App1 increased attachment (1.8-fold) and ingestion (3.4-fold) of yeast cells by AMs as compared with the WT strain (P = 0.01). Reintroduction of the APP1 gene (Δapp1Rec) restored the WT phenotype (Figure 4a). Therefore, App1 is a novel cryptococcal protein that exerts a unique and specific function in the regulation of phagocytosis of C. neoformans.
Figure 4.
App1 regulates phagocytosis of C. neoformans. (a and b) Absence of App1 (Δapp1) increases attached and ingested cells by AMs. Reconstitution of APP1 gene (Δapp1Rec) as well as treatment with App1 recombinant protein restores the WT phenotype (a) as compared with the control (b). (c) App1 treatment decreases phagocytosis of C. neoformans WT cells. (d) Ex vivo phagocytosis. Downregulation of Ipc1 and absence of App1 increase phagocytosis of fungal cells by AMs after 2 hours of infection as compared with the WT strain. IPC1Rec and Δapp1Rec strains reconstitute the WT phenotype. (e) App1 does not inhibit phagocytosis of antibody-coated erythrocytes. (f) Treatment with App1 inhibits phagocytosis of iC3b/IgM-IgG–coated erythrocytes. WT, WT C. neoformans H99 strain; Δapp1, C. neoformans lacking App1 protein; Δapp1Rec, Δapp1 reconstituted strain; IPC1Rec, Ipc1 reconstituted strain; App1 1×, 8 ng of App1 recombinant protein plus 14 ng of contaminating proteins; App1 2×, 3×, 4×, and 5×, two-, three-, four-, and fivefold App1 1× concentration; control 1×, 14 ng of contaminating proteins; control 2×, 3×, 4×, and 5×, two-, three-, four-, and fivefold control 1× concentration.
Production of App1 recombinant proteins and effect of App1 treatment on phagocytosis of C. neoformans.
Since the above studies suggested a role for App1 in phagocytosis, it became important to determine the effect(s) of the protein on phagocytosis directly. App1 recombinant protein was therefore produced by expressing C. neoformans App1 cDNA from serotype A strain H99 into a prokaryotic expression system, as described in Methods. Clarified cell lysate obtained from the expression of pBAD-His-App1 was first loaded to a HiTrap chelating HP column loaded with nickel and then to a HiTrap Q chromatography column. Fractions from each column were analyzed by Western blotting and silver staining (data not shown), and the fraction with the highest ratio of App1 to contaminating proteins (Q54) was used for cellular treatments. The lysate from control cells expressing pBAD-His (mock) vector was subjected to an identical purification protocol, and the same fraction collected from the App1 purification was also collected from the control lysate. Silver staining analysis confirmed that the pattern of residual contaminating proteins in the purified App1 preparation used for cellular treatments was identical to the protein pattern in the respective control sample (data not shown). The concentration of App1 was determined by the difference in the protein concentration between the fraction containing App1 and the one obtained from control cells. As can be seen from Figure 4a, addition of App1 significantly decreased phagocytosis of the Δapp1 strain in a dose-dependent manner, whereas no difference in phagocytosis was observed when the control fraction was used at the same concentration (Figure 4b). This antiphagocytic effect was also observed when C. neoformans WT strain H99 was tested. Indeed, as for the Δapp1 strain, addition of App1 decreased both attachment and ingestion of the WT strain in a dose-dependent manner, whereas treatment with the control preparation showed no effect on phagocytosis (Figure 4c). Therefore, by using both genetic and pharmacological approaches, these results indicate that App1 exerts a novel function in the regulation of phagocytosis of C. neoformans by AMs.
We also tested whether the addition of App1 would inhibit phagocytosis of other yeast cells, such as Candida albicans. We found that App1 inhibited phagocytosis of C. albicans by affecting both attachment and ingestion (data not shown). Finally, we tested whether addition of App1 would have a toxic effect on AMs. After pharmacological treatment, cellular toxicity was evaluated by adding trypan blue to the AMs. We found no significant difference on the viability of AMs between App1- versus control-treated cells (data not shown).
Analysis of phagocytosis of C. neoformans WT H99, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains by AMs was also performed ex vivo. Mice were infected intranasally with the above strains, and two hours after inoculation AMs were collected by BAL, and phagocytic indexes were determined by microscopic observation. As expected, we found that the ex vivo phagocytosis is more efficient than the in vitro phagocytosis (more yeast cells are found attached or ingested by AMs in a 2 hour period). Importantly, however, the difference in phagocytic indexes among the strains obtained ex vivo was similar to that obtained in vitro. Indeed, the phagocytic indexes for attached and ingested yeast cells for GAL7:IPC1 were increased by 1.9- and 2.4-fold, respectively, and the phagocytic index for attached and ingested yeast cells for Δapp1 were increased by 2.1- and 3.1-fold, respectively, as compared with the WT H99 strain (Figure 4d). This difference in phagocytosis indexes is similar to that described in Figure 1 for the GAL7:IPC1-glucose strain and in Figure 4a for the Δapp1 strain. We found no significant differences among the ex vivo phagocytic indexes of WT H99, IPC1Rec, and Δapp1Rec strains (Figure 4d).
To investigate the molecular mechanism(s) by which App1 inhibits phagocytosis, we tested whether App1 would inhibit phagocytosis of antibody-coated erythrocytes and complement/antibody-coated erythrocytes. We found that whereas App1 did not inhibit attachment or ingestion of IgM/IgG-coated erythrocytes (Figure 4e), it did significantly inhibit both attachment and ingestion of iC3b-IgM/IgG–coated erythrocytes (Figure 4f) in a dose-dependent manner (one to four times). Most interestingly, phagocytosis of iC3b-IgM/IgG–coated erythrocytes was not further inhibited by addition of the highest concentration of App1 (five times) (Figure 4f). These results clearly suggest that App1 blocks the complement- and not the antibody-mediated phagocytosis of the iC3b-IgM/IgG–coated erythrocytes.
Phagocytosis assay and App1 pharmacological treatment were performed at least twice and using AMs isolated from two mouse models (see Methods). No significant difference was found in the phagocytic index between AMs collected from either mouse model. The data presented are the geometric means ± SDs of phagocytic indexes using AMs from Tgε26 mice.
Ipc1-App1 pathway regulates virulence in two different immunodeficient murine animal models.
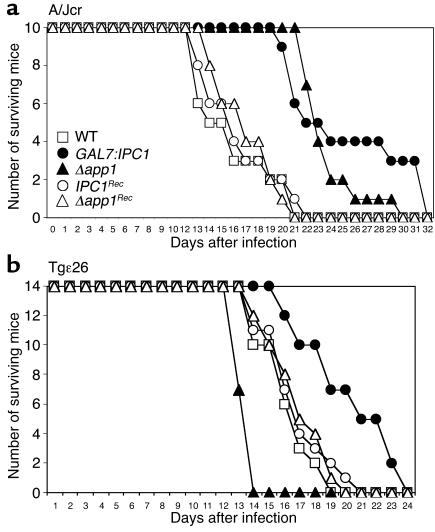
Since both Ipc1 and App1 inhibit phagocytosis of C. neoformans by AMs, we investigated whether they regulate pathogenicity of C. neoformans in a murine animal model immunodeficient for complement C5 (A/Jcr), when the lung is used as the port of entry. Mice were infected intranasally with C. neoformans WT H99, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains, and survival of the mice was monitored. The App1 protein is absent in the Δapp1 strain (Figure 3), whereas Ipc1 activity is indeed downregulated in the animal (data not shown), since glucose, instead of galactose, is available as a carbon source. The average survivals of GAL7:IPC1- and Δapp1-infected mice were 25.4 ± 5.2 and 21.7 ± 2.11 days, as compared with 14.9 ± 3.31 days for WT-infected mice (P = 0.002 and P = 0.01, respectively) (Figure 5a). Interestingly, survival of Δapp1-infected mice significantly decreased at a later phase of infection (26th day), whereas mice infected with the GAL7:IPC1 strain showed increased survival (40% GAL7:IPC1 versus 5% Δapp1, respectively). The IPC1Rec and Δapp1Rec strains showed average survivals of 18.8 ± 3.2 days and 17.1 ± 2.5 days, respectively, which are not significantly different from the average survival observed with WT-infected mice (P > 0.05). These results indicate that downregulation of Ipc1 or absence of App1 produces a strain less virulent than the WT in this mouse model.
Figure 5.
Virulence of C. neoformans in animal models. (a) Survival of A/Jcr mice infected with C. neoformans WT, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains; average mouse survivals were 14.9 ± 3.31, 25.4 ± 5.52, 21.7 ± 2.11, 18.8 ± 3.2, and 17.1 ± 2.5 days, respectively. The GAL7:IPC1- and Δapp1-infected mice lived significantly longer than WT mice (P = 0.002 and 0.01, respectively). (b) Survival of Tgε26 mice. Mice infected with the GAL7:IPC1 strain 10 lived significantly longer than WT-infected mice (20.14 ± 2.95 and 16.21 ± 1.76 days on average, respectively; P = 0.01), and mice infected with the Δapp1 strain showed a significantly decreased survival as compared with WT-infected mice (13.5 ± 0.51 versus 16.21 ± 1.76 days, respectively; P = 0.03). Mice infected with IPC1Rec and Δapp1Rec showed average survivals of 17 ± 2.2 and 15.85 ± 1.79 days, respectively, which were not significantly different from the average survival observed in WT-infected mice (P > 0.05).
Since infection by C. neoformans mainly occurs in immunocompromised hosts, such as patients with AIDS in whom T and NK cell number and function are significantly impaired (31–33), we investigated whether Ipc1 and App1 would have a role in the pathogenicity of C. neoformans in a murine model immunodeficient for T and NK cells. For this experiment, we used Tgε26 mice, which lack functional T and NK cells, mimicking the condition found in patients with AIDS (19). We infected Tgε26 mice intranasally and followed their survival. The average survival of GAL7:IPC1-infected Tgε26 mice was 20.14 ± 2.95 days, which was significantly longer than the 16.21 ± 1.76 days for H99-infected mice (P = 0.01) (Figure 5b). Surprisingly, Tgε26 mice infected with the Δapp1 mutant strain showed decreased survival (13.5 ± 0.51) as compared with the WT H99 strain (P = 0.03) (Figure 5b). The average survivals of IPC1Rec- and Δapp1Rec-infected mice were 17 ± 2.2 and 15.85 ± 1.79 days, similar to the average survival of 16.21 ± 1.76 days observed with the WT-infected mice (P > 0.05). These results indicate that the absence of App1 produces a strain more pathogenic than the WT in mice with severe NK and T cell immunodeficiencies.
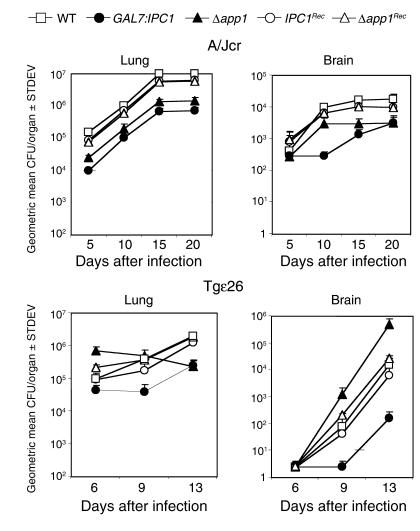
To determine whether the dissemination of yeast cells from the lung to the brain was affected by downregulation of Ipc1, by the absence of App1, and/or by the immune response of the host, lungs and brains were removed and assayed for viable C. neoformans. The number of GAL7:IPC1 and Δapp1 cells recovered from the lungs of A/Jcr-immunodeficient mice was significantly lower than the number of recovered WT cells (P = 0.02). Additionally, both GAL7:IPC1 and Δapp1 strains showed a decreased dissemination to the brain as compared with the WT strain, which was statistically significant at days 15 and 20 of infection (P = 0.01 and P = 0.015, respectively) (Figure 6, top panels). Therefore, Ipc1 and App1 are important for C. neoformans to establish infection in the brain of an A/Jcr-immunodeficient mouse model.
Figure 6.
Tissue-burden cultures of C. neoformans in animal models. Number of yeast cells expressed as geometric mean of CFU per organ recovered from lung (left panels) and brain (right panels) of A/Jcr (top panels) and Tgε26 (bottom panels) mice during infection (in days) with C. neoformans WT, GAL7:IPC1 10, Δapp1 31, IPC1Rec, and Δapp1Rec strains. Mice were infected intranasally, organs from five animals for each group were extracted and homogenized, and serial dilutions were plated for tissue-burden cultures at selected days after inoculation. Mice infected with the GAL7:IPC1 strain showed a significant decrease of yeast cell numbers in the brains of both animal models as compared with the WT strain (P = 0.01 and P = 0.001, respectively). Brains of A/Jcr mice infected with the Δapp1 strain showed a decreased concentration of yeast cells as compared with those infected with the WT strain (P = 0.015). In contrast, the brains of Tgε26 mice infected with the Δapp1 strain showed an increased number of yeast cells as compared with the brains infected with the WT strain (P = 0.002). Both IPC1Rec and Δapp1Rec strains reconstitute the phenotype observed with the WT H99 strain in both animal models.
In Tgε26-immunocompromised mice, however, the absence of App1 produced a strain that proliferated at a higher rate in the lung and disseminated readily to the brain as compared with the WT strain (P = 0.002) (Figure 6, bottom panels). Conversely, we found that Tgε26 mice infected with the GAL7:IPC1 strain showed a drastic decrease of fungal dissemination to the brain as compared with those infected with the WT strain (P = 0.001) (Figure 6, bottom panels). As expected, these data obtained with tissue-burden culture studies corroborate those obtained with survival studies, suggesting that absence of App1 produces a strain more pathogenic than the WT in mice with severe immunodeficiency, and that Ipc1 may regulate pathogenicity of C. neoformans through mechanisms other than modulation of App1. Both survival and tissue-burden culture studies were performed by using a second and independent GAL7:IPC1 13 and Δapp1 6 transformant, and no significant difference was found versus the corresponding previously used mutant strain (data not shown). Additionally, both IPC1Rec- and Δapp1Rec-infected mice showed survival and fungal cell dissemination similar to those observed with the WT-infected mice (Figures 5 and 6), showing that the WT phenotype(s) have been restored by reintroducing the gene(s) back into the corresponding knockout mutants.
Histopathology.
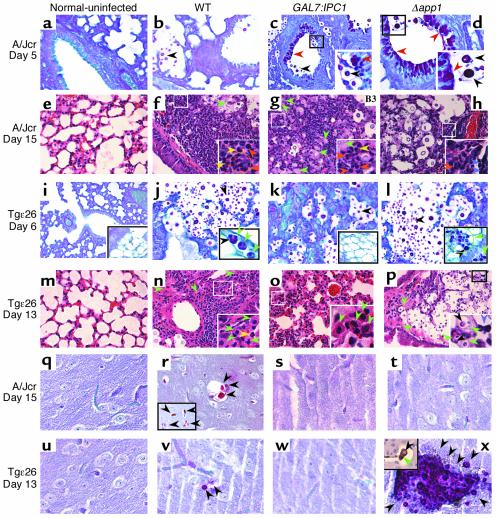
To analyze the effect of downregulation of Ipc1 and/or loss of App1 on the host inflammatory response in vivo, we studied histology of lungs and brains during the infection. The immunodeficient mouse models were infected with C. neoformans WT H99, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains, and at days 5 and 15 after inoculation for the A/Jcr mouse model or days 6 and 13 after inoculation for the Tgε26 mouse model, lungs and brains were collected, fixed in 10% formalin, and embedded in paraffin, and sections were stained with hematoxylin and eosin to visualize the host inflammatory response, PAS to identify the fungal cell wall and body, and mucicarmine (data not shown) as additional staining specific for the C. neoformans capsule (34). Selected sections were also subjected to immunohistochemical analysis using anti-CD68 antibody to identify macrophages.
In the mouse model with C5 deficiency (A/Jcr), an intense PAS-positive deposition due to mucus hyperplasia/metaplasia (stained red by PAS) at day 5 of the infection in bronchi and terminal bronchioles was found in lungs infected with GAL7:IPC1 (Figure 7c) or Δapp1 (Figure 7d) strains. Interestingly, this phenomenon was not present in bronchi and bronchioles not containing yeast cells or in lungs infected with the WT strain (Figure 7b), normal uninfected lung (Figure 7a), or IPC1Rec- and Δapp1Rec-infected mice (data not shown). The mucus hyperplasia/metaplasia was still detectable at day 15 of infection, although it was much less extensive (data not shown). At day 15 of the infection, the host cellular response was dramatically activated. In particular, the inflammatory response against the WT strain was comprised of polymorphonuclear cell infiltrates, such as neutrophils (orange arrowheads), with few eosinophils (yellow arrowheads), lymphocytes, and macrophages (green arrowhead, Figure 7f). Interestingly, the host cellular infiltrates in lung infected with the GAL7:IPC1 strain were also represented by neutrophils but with a large majority of macrophages and few lymphocytes and eosinophils (Figure 7g). Infiltrates in lungs infected with the Δapp1 strain also contained neutrophils but with a large number of lymphocyte infiltrations (violet arrowheads), with few macrophages and eosinophils (Figure 7h). Lung infected with either IPC1Rec or Δapp1Rec strains showed a cellular infiltration similar to that observed with the WT strain (data not shown).
Figure 7.
Histopathology of lungs and brains from animal models. (a–d, i–l, q–t, and u–x) PAS-positive staining. (e–h and m–p) Hematoxylin and eosin staining. Histopathology of lung from A/Jcr (a–d and e–h) and Tgε26 (i–l and m–p) is shown. Normal lung (a, e, i, and m) and lungs infected with C. neoformans WT (b, f, j, and n), GAL7:IPC1 (c, g, k, and o), and Δapp1 (d, h, l, and p) at 5 (a–d) and 15 (e–h) days and at 6 (i–l) and 13 (m–p) days after infection are shown. In the A/Jcr mouse model, a mucus hyperplasia/metaplasia (PAS-positive material) is present in lungs infected with the GAL7:IPC1 (c) and Δapp1 (d) strains (red arrowheads, c and d), but not in the lung infected with the WT strain (b). At day 15 of infection (e–h), intense neutrophil recruitment is observed in lung infected with the WT strain (orange arrowheads and orange circle, f), whereas neutrophils and macrophages are found in lung infected with the GAL7:IPC1 strain (orange arrowheads and green arrowheads, g) or neutrophils and lymphocytes are found in lung infected with the Δapp1 strain (orange arrowhead and violet arrowheads, h). At day 6 of infection in the Tgε26 mouse model (i–l), no mucus hyperproduction was observed (j–l and data not shown). Instead, dissemination of C. neoformans within macrophages in brown fat and lymphoid tissue was observed with the WT strain (green arrowhead, j) and, more pronounced, with the Δapp1 strain (green arrowhead and green circle, l) but not with the GAL7:IPC1 strain (k). At day 13 of infection (m–p), the inflammatory response against WT, GAL7:IPC1, and Δapp1 is mainly represented by macrophages (green arrowheads, n–p). The Δapp1 yeast cells are found mainly in brown fat (p). Histopathology of brains from A/Jcr (q–t) and Tgε26 (u–x) mice at days 15 and 13 of infection, respectively. Normal brains (q and u) and brains infected with WT (r and v), GAL7:IPC1 (s and w), and Δapp1 (t and x) are shown. C. neoformans cells were found in brains infected with the WT strain (r and v), whereas no yeast cells were found in brain sections examined from mice infected with the GAL7:IPC1 strain (s and w). The Δapp1 yeast cells were not found in brain sections examined from A/Jcr brain (t), whereas numerous Δapp1 cells were found in Tgε26 infected brain (x). Additionally, an Δapp1 yeast cell was found inside a macrophage (stained brown with anti-CD68 antibody) in a blood vessel of brain tissue at day 6 of infection (inset in x). (j, l, p, and x) In the insets, black arrowheads indicate yeast cells inside macrophages. (f and g) Yellow arrowheads indicate eosinophils. (c, d, f–h, and n–p) Insets are magnifications of small squared area. (r) Inset represents PAS staining of a different brain section. (x) Inset represents immunohistochemistry with anti-CD68 antibody for macrophage staining. Histology studies were also performed using IPC1Rec and Δapp1Rec strains, showing a phenotype similar to that observed with the WT strain (data not shown). Magnification, ×40. The images are representative of three different organ sections.
In the mouse model with impaired T and NK cells (Tgε26), no mucus hyperplasia/metaplasia was found at day 6 of the infection in lungs infected with either WT (Figure 7j), GAL7:IPC1 (Figure 7k), Δapp1 (Figure 7l, IPC1Rec, or Δapp1Rec strains (data not shown). Interestingly, a local dissemination of WT fungal cells inside macrophages was found in lymphatic vessel and fat tissue in proximity of the lung (green arrowheads in inset of Figure 7j). This phenomenon was more evident with the Δapp1 mutant, and macrophages, loaded with this mutant strain, infiltrated not only fat tissue but also pulmonary lymph nodes (green arrowheads in inset of Figure 7l). This local dissemination was not observed in lung sections infected with the GAL7:IPC1 strain (Figure 7k). In contrast to the A/Jcr model, at day 13 of infection the inflammatory response in the Tgε26 mouse model against the WT strain was comprised mostly of macrophages with few lymphocytes, whereas neutrophils were absent in the cellular infiltrates (Figure 7n). The recruitment of macrophages was more evident in lungs infected with the GAL7:IPC1 strain, where an intense interstitial infiltration of macrophages represented the major host inflammatory response against this strain (Figure 7o). Interestingly, in lung infected with Δapp1 strain, a dramatic dissemination of fungal cells in fat tissue and local lymph nodes was found. As shown in Figure 7p, fat tissue and residual lymphoid tissue between two bronchi (upper left and lower left) were completely replaced by C. neoformans Δapp1 cells.
Histology of brain tissues from both animal models infected with WT, GAL7:IPC1, and Δapp1 strains corroborated the results obtained with tissue-burden studies. Animals infected with GAL7:IPC1 strain showed a decreased fungal dissemination as compared with the WT strain in both animal models (Figure 7, s and w versus r and v). Animals infected with Δapp1 mutant showed a decreased dissemination in the A/Jcr model (Figure 7t), whereas in Tgε26, the mutant strain reached the brain as early as 6 days after infection (inset in Figure 7x) and more dramatically at 13 days (Figure 7x).
These results clearly show that the host lung inflammatory response against C. neoformans between the A/Jcr and Tgε26 mouse models is different, with a prevalent neutrophil infiltration in the A/Jcr model and a prevalent macrophage infiltration in the Tgε26 model. These studies indicate that whereas dissemination of the GAL7:IPC1 strain is impaired, the Δapp1 strain readily disseminates within macrophages to fat and lymphoid tissues found in the mediastinum area. Histology of lung and brain was performed using two animals for each strain (WT, GAL7:IPC1, Δapp1, IPC1Rec, and Δapp1Rec strains). No significant differences were found between the animals infected with the same strain. The cellular infiltrates observed with IPC1Rec and Δapp1Rec strains were similar to those observed with the WT strain (data not shown). Macrophage involvement was demonstrated using anti-CD68 antibody and performed in all tissue sections examined (Figure 7x and data not shown).
Discussion
We previously demonstrated that regulation of Ipc1 plays a major role in pathogenicity of C. neoformans in the rabbit animal model (9). This model allows the study of the impact of a gene of interest on pathogenicity of C. neoformans during the late stage of cryptococcosis (meningoencephalitis). In the present study, we investigated the role of Ipc1 during early stages of the infection using murine animal models of cryptococcosis. Since the lung is the port of entry for C. neoformans, AMs are one of the first host defenses against the inhaled microorganism. Indeed, we found that Ipc1 regulates phagocytosis of C. neoformans by AMs, and this regulation occurs through the modulation of a newly identified protein, App1.
During the 1980s and 1990s, we had focused on the identification and purification of cytoplasmic factor(s) of C. neoformans involved in the regulation of phagocytosis of yeast cells by AMs. These early studies resulted in the partial isolation and purification, from crude cytoplasmic extract, of an approximately 20-kDa protein as a cryptococcal factor that specifically inhibits phagocytosis (29). Rabbit polyclonal antibodies were raised against this cytoplasmic factor and used to screen a C. neoformans cDNA library. A putative gene encoding for an approximately 20-kDa protein was identified and sequenced from both serotypes A and D of C. neoformans (unpublished observations; GenBank accession numbers AF180107, AF180108, and AY101600). Using polyclonal antibodies, it was shown that this cytoplasmic factor is secreted extracellularly in culture medium of C. neoformans (29) (Figure 3). These results were corroborated by the fact that this protein was found in serum of patients with AIDS who had disseminated cryptococcosis (Figure 3d) (35). In this study, we isolated by DD-RT-PCR the same gene isolated previously, whose expression is regulated by the level of Ipc1 activity. This gene was named APP1 for antiphagocytic protein 1, and through a genetic, biochemical, and immunological approach we characterized the function of App1 on phagocytosis and virulence of C. neoformans.
The APP1 gene appears to be regulated at the transcriptional level by modulation of Ipc1 activity (Figure 2b). In addition, DD-RT-PCR identified three other fragments to be regulated by IPC1 expression (data not shown), but no homology to any other gene sequences was found. We focused our attention on App1 (Figure 2a) because it was already isolated and shown to exert a specific function on the phagocytosis of C. neoformans. Since Ipc1 transcriptionally modulates the expression of APP1, it is likely that this regulation occurs at the promoter region of the APP1 gene, although Ipc1 may also regulate the stability of APP1 mRNA. A search of putative transcription factors present in the APP1 promoter (Web promoter scan service provided by the BioInformatics and Molecular Analysis Section at NIH, http://molbio.info.nih.gov/molbio/db.html) showed significant hits for the following transcription factors: AP-2, CREB, and ATF. In our studies, we found that modulation of Ipc1 regulates the level of diacylglycerol (DAG) (unpublished observation) (36). Interestingly, it has been shown that DAG may exert a regulatory effect on the transcription factor ATF/cAMP-responsive element in RINm5F cells (37). Since APP1 promoter contains this putative signal, it is reasonable to hypothesize that transcriptional activation of APP1 is mediated through the lipid regulated by Ipc1.
Because the native App1 protein previously isolated was involved in the inhibition of phagocytosis, we disrupted the APP1 gene in C. neoformans and studied whether phagocytosis would be affected by the absence of App1. We found that, indeed, phagocytosis of C. neoformans increased when App1 protein was absent (Figure 4a). Since App1 does not affect other cryptococcal factors known to influence phagocytosis, such as the capsule, it is proposed that App1 regulates phagocytosis through a specific and novel molecular mechanism. Addition of recombinant App1 protein or reconstitution of APP1 gene in the Δapp1 strain restores the Δapp1 phenotype nearly at the WT level (Figure 4a). App1 treatment further reduces phagocytosis of the WT strain (Figure 4c) by decreasing both attachment and ingestion of yeast cells by AMs, although the inhibitory effect of App1 seems more pronounced on the ingestion process (Figure 4c). The observation that App1 also inhibits phagocytosis of C. albicans (data not shown) suggests that App1 may act by blocking a common receptor responsible for attachment and internalization of these yeasts. The fact that App1 does not block phagocytosis of antibody-coated erythrocytes, whereas it does inhibit the phagocytosis of complement/antibody-coated erythrocytes, clearly suggests that App1 exerts its effect through the inhibition of complement-mediated phagocytosis, which is a common molecular mechanism by which fungal cells are phagocytosed. Indeed, several studies have suggested that phagocytosis of C. neoformans is primarily mediated through complement receptors (CR1, CR3, and CR4), although other mechanisms are also involved, such as antibody-mediated (anti-GXM) and mannose-mediated mechanisms (38–41). Among the complement receptors, CR3 (CD11b/CD18) plays a major role in the attachment and internalization of C. albicans (42), Blastomyces dermatitidis (43), C. neoformans (40, 44, 45), and bacteria (46) by the host’s phagocytic cells. Since our results show that App1 blocks iC3b-mediated phagocytosis, it is reasonable to hypothesize that App1 inhibits the internalization of C. neoformans by blocking the interaction of fungal cells with the complement receptor. It remains to be established which complement receptor(s) are most affected by App1 action.
The provirulence effects of Ipc1 are mediated through the regulation of various factors, such as melanin formation, intracellular growth (9), and phagocytosis through App1 (this study). Differential modulation of these factors by Ipc1 could be exerted at different stages of the infection (i.e., initiation, dormancy, reactivation, dissemination, proliferation). Due to its antiphagocytic role, App1 may be produced by C. neoformans during early stages of the infection (initiation-dissemination). Once inhaled, C. neoformans cells are phagocytosed by AMs, and this internalization is required for effective killing of the microorganism, when the host cellular response is functional. In a mouse model deficient for complement C5, in which the immune response is still cell mediated, we found that downregulation of Ipc1 and absence of App1 decrease virulence. However, C. neoformans is a fungal pathogen that primarily afflicts immunocompromised subjects such as those affected by HIV, where the cell-mediated immune response (T and NK) is significantly impaired. In these patients, phagocytosis might not represent an effective mechanism of defense by the host, when the activation of T and/or NK cells and, thus, the production of activated macrophages are significantly reduced (6, 47–49). Moreover, in the case of facultative intracellular pathogens, such as C. neoformans, for which intracellular growth can be a favorable condition, phagocytosis may represent a successful strategy for better survival in the host. Indeed, when we investigated the effect of absence of App1 in an immunocompromised mouse model deficient for T and NK cells, we found that virulence (Figure 5b) and dissemination of C. neoformans infection were significantly enhanced (Figure 6, bottom panels). As a result, absence of App1 protein significantly increases the number of yeast cells recovered from the brain. These results indicate that lack of App1 favors the internalization of C. neoformans into nonactivated macrophages and the dissemination of fungal cells to the brain when T and NK cells are not functional.
The host inflammatory response to the C. neoformans infection in A/Jcr and Tgε26 mice is different. First, in the A/Jcr model, a mucus hyperplasia/metaplasia is present in bronchi and bronchioles of the lung infected by GAL7:IPC1 or Δapp1 strains (Figure 7, c and d), and it seems to be stimulated by the nearby presence of the GAL7:IPC1 or Δapp1 cells, since it is absent in airways that do not contain yeast cells. It has been reported that mucus contains mucin (MUC7) and histatin-5 (Hsn-5), two peptides that, among their spectrum of antimicrobial activity, display a potent killing effect against a variety of fungi, including C. albicans and C. neoformans (50–53). This hyperproduction of mucus may represent a host defense mechanism against inhaled microorganisms, and, interestingly, it appears to be stimulated particularly by less virulent strains, since it is less evident in lung infected with the WT strain (Figure 7b). In recent years, studies have shown that activation of the Th2 response leads to mucus hypersecretion (54). The A/J mice, and presumably also the A/Jcr mice, are prone to develop a Th2 response due to the downregulation of IL-12 in the absence of C5 (26–28). This could explain why the mucus hyperplasia/metaplasia is observed in the A/Jcr model and not in the Tgε26 mice. However, it does not completely explain why the mucus hypersecretion is not stimulated in the A/Jcr mice infected with the WT strain, suggesting that this phenomenon is a complex process, also determined by the characteristics of the pathogen(s). Second, the cellular inflammatory response in the A/Jcr mouse model is mainly represented by neutrophils, whereas in the Tgε26 mouse model it is mainly represented by macrophages (Figure 7; compare f, g, and h with n, o, and p, respectively). This difference in the inflammatory response may account for the difference we observed in virulence among the WT, GAL7:IPC1, and Δapp1 strains, especially in the Tgε26 mice. Although macrophages are taking up more GAL7:IPC1 cells than the WT, this mutant strain shows a profound defect in intracellular growth (due to its defect on growth at pH 4.0) and on melanin production (9). In this case, a macrophage response may be able to partially control the GAL7:IPC1 dissemination. In the case of infection by Δapp1, however, the macrophage response in Tgε26 mice is clearly unable to control the dissemination. The Δapp1 strain is more internalized by macrophages than the GAL7:IPC1 mutant; it does not have a defect on growth at low pH or an alteration of melanin production. Its dissemination seems to be determined by the host’s immune response. In a mouse model deficient for C5 (A/Jcr), macrophages and lymphocytes are recruited into the lung decreasing fungal dissemination to the brain (Figures 6 and 7t). In contrast, in a mouse model deficient for T and NK (Tgε26), the dissemination of the Δapp1 strain occurs very early during the infection (day 6) into extrapulmonary tissue (Figure 7, l and p) and into the brain (inset in Figure 7x). Lack of App1 seems also to favor recruitment of lymphocytes in the lung of A/Jcr mice (Figure 7h) as compared with the WT (Figure 7f), suggesting that App1 may exert an inhibitory effect on other cells involved in the host cellular immunity, such as lymphocytes.
The physiological mechanism by which this dissemination occurs remains to be elucidated. However, these studies clearly suggest that, without App1, C. neoformans is more easily taken up by macrophages, which, in mice with an intact T and NK cellular response, can control/kill the yeast. In T-NK–deficient mice, the macrophage may allow the yeast to be “protected” as it migrates from the lung to the brain. In other words, the macrophage can act as a “Trojan horse” for the yeast to reach other tissues (Figure 7l) and the central nervous system (Figure 7x), and this mechanism of dissemination is also observed with the WT strain (Figure 7j) and in previously reported observations (55). On the other hand, impairment of a cellular response could favor the yeast growth inside the macrophages, inducing their lysis. The released yeast cells in the extracellular matrix can then reach the central nervous system through the blood stream. Further studies are needed to investigate these potential mechanisms.
In the same immunocompromised conditions (Tgε26), downregulation of Ipc1 resulted in a dramatic reduction of the dissemination of C. neoformans cells to the brain (Figure 6, bottom) or other tissues (Figure 7k), suggesting that the effect of the modulation of Ipc1 on virulence of C. neoformans is not only mediated through the regulation of App1 and phagocytosis. This hypothesis is further supported by the results obtained in the A/Jcr model in which the antipathogenic effect obtained by downregulating Ipc1 was more pronounced than that observed when App1 protein was absent. Indeed, we previously demonstrated that Ipc1 regulates other downstream effects, such as melanin production and intracellular growth, and these factors are important for the outcome of the infection (9, 36, 56, 57).
In conclusion, this study defines a novel function for Ipc1 in the regulation of pathogenicity of C. neoformans during early stages of the infection. In particular, we identify that modulation of Ipc1 regulates the level of a novel protein, App1. Biochemical characterization of App1 reveals its function as an antiphagocytic factor. The Ipc1-App1 pathway regulates phagocytosis of C. neoformans by AMs and virulence in the murine animal models of disseminated cryptococcosis by potentially affecting the numbers of the intra- versus extracellular yeast cells. Importantly, we show that this trait assumes a crucial role for the development of cryptococcosis in immunocompromised hosts.
Acknowledgments
We are grateful to Stephen Tomlinson, Lina Obeid, and Stephanie Tucker for discussions. We thank Russell Harley for helping in the analysis of the histological sections. Special thanks go to Elizabeth Collins, Allyson Plowden, and Zainab Amani for technical assistance; Peter Nicholas, Emily Pauling, Lu Wang, and Anna Maria Porcelli for helping with Tgε26 mouse experiments; Angela Restrepo for providing sera of patients with AIDS and normal subjects; Margaret Romano for histology staining; and LuAnne Harley for helping on the preparation of this manuscript. This work was supported in part by NIH HL43707, NIH AI28388, NIHAI44975, MUCU Institutional Project 21363, URC 24374, and NIH RR17677-01 project 2 to M. Del Poeta from the Centers of Biomedical Research Excellence Program of the National Center for Research Resources. M. Del Poeta is a Burroughs Wellcome New Investigator in Pathogenesis of Infectious Diseases.
Footnotes
Chiara Luberto and Beatriz Martinez-Mariño contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: alveolar macrophages (AMs); antiphagocytic protein 1 (App1); inositol phosphoryl ceramide synthase 1 (Ipc1); yeast nitrogen base (YNB); yeast peptone dextrose (YPD); yeast extract peptone (YP); bronchoalveolar lavage (BAL); 5% nonfat milk/1× PBS/0.1% Tween 20 (5% NFM/PBST); 5′ untranslated region (5′-UTR); open reading frame (ORF); 3′ untranslated region (3′-UTR); periodic acid–Schiff (PAS); differential display RT-PCR (DD-RT-PCR); inhibitory protein 1 (IP1); diacylglycerol (DAG).
References
- 1.Casadevall, A., and Perfect, J.R. 1998. Cryptococcus neoformans.American Society for Microbiology. Washington, DC, USA. 407–456.
- 2.Diamond RD, May JE, Kane MA, Frank MM, Bennett JE. The role of the classical and alternate complement pathways in host defenses against Cryptococcus neoformans infection. J. Immunol. 1974;112:2260–2270. [PubMed] [Google Scholar]
- 3.Levitz, S.M. 1994. Macrophage-cryptococcus interactions. In Macrophage-pathogen interactions. Marcel Dekker Inc. New York, New York, USA. 533–543. [PubMed]
- 4.Murphy JW, Zhou A, Wong SC. Direct interactions of human natural killer cells with Cryptococcus neoformans inhibit granulocyte-macrophage colony-stimulating factor and tumor necrosis factor alpha production. Infect. Immun. 1997;65:4564–4571. doi: 10.1128/iai.65.11.4564-4571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawakami K, et al. NK cells eliminate Cryptococcus neoformans by potentiating the fungicidal activity of macrophages rather than by directly killing them upon stimulation with IL-12 and IL-18. Microbiol. Immunol. 2000;44:1043–1050. doi: 10.1111/j.1348-0421.2000.tb02601.x. [DOI] [PubMed] [Google Scholar]
- 6.Murphy JW, Hidore MR, Wong SC. Direct interactions of human lymphocytes with the yeast-like organism, Cryptococcus neoformans. J. Clin. Invest. 1993;91:1553–1566. doi: 10.1172/JCI116361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox GM, et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 8.Noverr MC, Toews GB, Huffnagle GB. Production of prostaglandins and leukotrienes by pathogenic fungi. Infect. Immun. 2002;70:400–402. doi: 10.1128/IAI.70.1.400-402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luberto C, et al. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 2001;15:201–212. doi: 10.1101/gad.856001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond RD, Bennett JE. Disseminated cryptococcosis in man: decreased lymphocyte transformation in response to Cryptococcus neoformans. J. Infect. Dis. 1973;127:694–697. doi: 10.1093/infdis/127.6.694. [DOI] [PubMed] [Google Scholar]
- 11.Feldmesser M, Tucker S, Casadevall A. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 2001;9:273–278. doi: 10.1016/s0966-842x(01)02035-2. [DOI] [PubMed] [Google Scholar]
- 12.Reardon CC, Kim SJ, Wagner RP, Kornfeld H. Interferon-gamma reduces the capacity of human alveolar macrophages to inhibit growth of Cryptococcus neoformans in vitro. Am. J. Respir. Cell Mol. Biol. 1996;15:711–715. doi: 10.1165/ajrcmb.15.6.8969264. [DOI] [PubMed] [Google Scholar]
- 13.Diamond RD, Root RK, Bennett JE. Factors influencing killing of Cryptococcus neoformans by human leukocytes in vitro. J. Infect. Dis. 1972;125:367–376. doi: 10.1093/infdis/125.4.367. [DOI] [PubMed] [Google Scholar]
- 14.Diamond RD, Bennett JE. Growth of Cryptococcus neoformans within human macrophages in vitro. Infect. Immun. 1973;7:231–236. doi: 10.1128/iai.7.2.231-236.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6) J. Immunol. 1998;160:2393–2400. [PubMed] [Google Scholar]
- 16.Chang WL, van der Heyde HC, Klein BS. Flow cytometric quantitation of yeast: a novel technique for use in animal model work and in vitro immunologic assays. J. Immunol. Methods. 1998;211:51–63. doi: 10.1016/s0022-1759(97)00191-9. [DOI] [PubMed] [Google Scholar]
- 17.Levitz SM, et al. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker SC, Casadevall A. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3165–3170. doi: 10.1073/pnas.052702799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, et al. A block in both early T lymphocyte and natural killer cell development in transgenic mice with high-copy numbers of the human CD3E gene. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9402–9406. doi: 10.1073/pnas.91.20.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu B, Sonstein J, Christensen PJ, Punturieri A, Curtis JL. Deficient in vitro and in vivo phagocytosis of apoptotic T cells by resident murine alveolar macrophages. J. Immunol. 2000;165:2124–2133. doi: 10.4049/jimmunol.165.4.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 9.31–9.58.
- 23.Cox GM, Toffaletti DL, Perfect JR. Dominant selection system for use in Cryptococcus neoformans. J. Med. Vet. Mycol. 1996;34:385–391. [PubMed] [Google Scholar]
- 24.Wang B, Simpson SJ, Hollander GA, Terhorst C. Development and function of T lymphocytes and natural killer cells after bone marrow transplantation of severely immunodeficient mice. Immunol. Rev. 1997;157:53–60. doi: 10.1111/j.1600-065x.1997.tb00973.x. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes JC, Wicker LS, Urba WJ. Genetic control of susceptibility to Cryptococcus neoformans in mice. Infect. Immun. 1980;29:494–499. doi: 10.1128/iai.29.2.494-499.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z, Stevenson MM. IL-12 is required for antibody-mediated protective immunity against blood-stage Plasmodium chabaudi AS malaria infection in mice. J. Immunol. 2002;168:1348–1355. doi: 10.4049/jimmunol.168.3.1348. [DOI] [PubMed] [Google Scholar]
- 27.Sam H, Stevenson MM. Early IL-12 p70, but not p40, production by splenic macrophages correlates with host resistance to blood-stage Plasmodium chabaudi AS malaria. Clin. Exp. Immunol. 1999;117:343–349. doi: 10.1046/j.1365-2249.1999.00966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp CL, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 2000;1:221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Mariño, B., Del Poeta, M., Perfect, J.R., and Bolaños, B. 1999. Cloning and expression of a Cryptococcus neoformans gene encoding a protein associated with inhibition of phagocytosis. In Abstracts of the 99th general meeting of the American Society for Microbiology. May 30–June 3. Chicago, Illinois, USA. 76.
- 30.Falquet L, et al. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratcliffe LT, Lukey PT, MacKenzie CR, Ress SR. Reduced NK activity correlates with active disease in HIV — patients with multidrug-resistant pulmonary tuberculosis. Clin. Exp. Immunol. 1994;97:373–379. doi: 10.1111/j.1365-2249.1994.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan RA, et al. Idiopathic CD4+ T-lymphocytopenia — four patients with opportunistic infections and no evidence of HIV infection. N. Engl. J. Med. 1993;328:393–398. doi: 10.1056/NEJM199302113280604. [DOI] [PubMed] [Google Scholar]
- 33.Fehniger TA, et al. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J. Immunol. 1998;161:6433–6438. [PubMed] [Google Scholar]
- 34.Lazcano O, et al. Combined histochemical stains in the differential diagnosis of Cryptococcus neoformans. Mod. Pathol. 1993;6:80–84. [PubMed] [Google Scholar]
- 35.Salgado, D.C., Lebrón de Jesús, J.M., and Bolaños, B. 1994. Purification and characterization of a cytoplasmic immunosuppressive component from Cryptococcus neoformans by preparative electrophoretic techniques. In Abstracts of the 94th general meeting of the American Society for Microbiology. March 23–27. Las Vegas, Nevada, USA. 77.
- 36.Luberto, C., Heung, L.J., Hannun, Y.A., and Del Poeta, M. 2002. Inositol phosphoryl ceramide synthase 1 (IPC1) plays a major role in the regulation of sphingolipid levels. In Abstracts of the 5th International Conference on Cryptococcus and Cryptococcosis. March 3–8. Adelaide, Australia. 180.
- 37.Welsh N. Interleukin-1 beta-induced ceramide and diacylglycerol generation may lead to activation of the c-Jun NH2-terminal kinase and the transcription factor ATF2 in the insulin-producing cell line RINm5F. J. Biol. Chem. 1996;271:8307–8312. doi: 10.1074/jbc.271.14.8307. [DOI] [PubMed] [Google Scholar]
- 38.Mansour MK, Levitz SM. Interactions of fungi with phagocytes. Curr. Opin. Microbiol. 2002;5:359–365. doi: 10.1016/s1369-5274(02)00342-9. [DOI] [PubMed] [Google Scholar]
- 39.He W, Casadevall A, Lee SC, Goldman DL. Phagocytic activity and monocyte chemotactic protein expression by pulmonary macrophages in persistent pulmonary cryptococcosis. Infect. Immun. 2003;71:930–936. doi: 10.1128/IAI.71.2.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taborda CP, Casadevall A. CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity. 2002;16:791–802. doi: 10.1016/s1074-7613(02)00328-x. [DOI] [PubMed] [Google Scholar]
- 41.Vecchiarelli A, Pietrella D, Bistoni F, Kozel TR, Casadevall A. Antibody to Cryptococcus neoformans capsular glucuronoxylomannan promotes expression of interleukin-12Rβ2 subunit on human T cells in vitro through effects mediated by antigen-presenting cells. Immunology. 2002;106:267–272. doi: 10.1046/j.1365-2567.2002.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsyth CB, Plow EF, Zhang L. Interaction of the fungal pathogen Candida albicans with integrin CD11b/CD18: recognition by the I domain is modulated by the lectin-like domain and the CD18 subunit. J. Immunol. 1998;161:6198–6205. [PubMed] [Google Scholar]
- 43.Newman SL, Chaturvedi S, Klein BS. The WI-1 antigen of Blastomyces dermatitidis yeasts mediates binding to human macrophage CD11b/CD18 (CR3) and CD14. J. Immunol. 1995;154:753–761. [PubMed] [Google Scholar]
- 44.Dong ZM, Murphy JW. Cryptococcal polysaccharides bind to CD18 on human neutrophils. Infect. Immun. 1997;65:557–563. doi: 10.1128/iai.65.2.557-563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross CE, Collins HL, Bancroft GJ. CR3-dependent phagocytosis by murine macrophages: different cytokines regulate ingestion of a defined CR3 ligand and complement-opsonized Cryptococcus neoformans. Immunology. 1997;91:289–296. doi: 10.1046/j.1365-2567.1997.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agramonte-Hevia J, Gonzalez-Arenas A, Barrera D, Velasco-Velazquez M. Gram-negative bacteria and phagocytic cell interaction mediated by complement receptor 3. FEMS Immunol. Med. Microbiol. 2002;34:255–266. doi: 10.1111/j.1574-695X.2002.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 47.Kawakami K, et al. IL-18 contributes to host resistance against infection with Cryptococcus neoformans in mice with defective IL-12 synthesis through induction of IFN-gamma production by NK cells. J. Immunol. 2000;165:941–947. doi: 10.4049/jimmunol.165.2.941. [DOI] [PubMed] [Google Scholar]
- 48.Salkowski CA, Balish E. Role of natural killer cells in resistance to systemic cryptococcosis. J. Leukoc. Biol. 1991;50:151–159. doi: 10.1002/jlb.50.2.151. [DOI] [PubMed] [Google Scholar]
- 49.Hidore MR, Nabavi N, Sonleitner F, Murphy JW. Murine natural killer cells are fungicidal to Cryptococcus neoformans. Infect. Immun. 1991;59:1747–1754. doi: 10.1128/iai.59.5.1747-1754.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bobek LA, Situ H. MUC7 20-Mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 2003;47:643–652. doi: 10.1128/AAC.47.2.643-652.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satyanarayana J, et al. Divergent solid-phase synthesis and candidacidal activity of MUC7 D1, a 51-residue histidine-rich N-terminal domain of human salivary mucin MUC7. J. Pept. Res. 2000;56:275–282. doi: 10.1034/j.1399-3011.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- 52.Situ H, Bobek LA. In vitro assessment of antifungal therapeutic potential of salivary histatin-5, two variants of histatin-5, and salivary mucin (MUC7) domain 1. Antimicrob. Agents Chemother. 2000;44:1485–1493. doi: 10.1128/aac.44.6.1485-1493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gururaja TL, et al. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7)1. . Biochim. Biophys. Acta. 1999; 1431:107–119. doi: 10.1016/s0167-4838(99)00034-5. [DOI] [PubMed] [Google Scholar]
- 54.Fahy JV. Goblet cell and mucin gene abnormalities in asthma. Chest. 2002;122:320S–326S. doi: 10.1378/chest.122.6_suppl.320s. [DOI] [PubMed] [Google Scholar]
- 55.Chretien F, et al. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J. Infect. Dis. 2002;186:522–530. doi: 10.1086/341564. [DOI] [PubMed] [Google Scholar]
- 56.Luberto, C., et al. 2002. Role of inositol phosphoryl ceramide synthase 1 (IPC1) in the regulation of phagocytosis. In Abstracts of the 5th International Conference on Cryptococcus and Cryptococcosis. March 3–8. Adelaide, Australia. 181.
- 57.Heung, L.J., Plowden, A., Luberto, C., and Del Poeta, M. 2002. Role of protein kinase C (PKC1) gene from Cryptococcus neoformans in the regulation of melanin production. In Abstracts of the 5th International Conference on Cryptococcus and Cryptococcosis. March 3–8. Adelaide, Australia.171.