Abstract
The rack-induced bonding mechanism of metals to proteins is a useful concept for explaining the generation of metal sites in electron transfer proteins, such as the blue copper proteins, that are designed for rapid electron transfer. The trigonal pyramidal structure imposed by the protein with three strong equatorial ligands (one Cys and two His) provides a favorable geometry for both cuprous and cupric oxidation states. However, the crystal structures of the Met121His mutant of azurin from Alcaligenes denitrificans at pH 6.5 (1.89- and 1.91-Å resolutions) and pH 3.5 (2.45-Å resolution) show that the preformed metal binding cavity in the protein is more flexible than expected. At high pH (6.5), the Cu site retains the same three equatorial ligands as in the wild-type azurin and adds His121 as a fourth strong ligand, creating a tetrahedral copper site geometry with a green color referred to as 1.5 type. In the low pH (3.5) structure, the protonation of His121 causes a conformational change in residues 117–123, moving His121 away from the copper. The empty coordination site is occupied by an oxygen atom of a nitrate molecule of the buffer solution. This axial ligand is coordinated less strongly, generating a distorted tetrahedral copper geometry with a blue color and spectroscopic properties of a type-1 site. These crystal structures demonstrate that blue copper proteins are flexible enough to permit a range of movement of the Cu atom along the axial direction of the trigonal pyramid.
Keywords: x-ray crystallography, copper protein
It has been a long-standing dogma in the field of metalloprotein research that the metal site in a metalloprotein is configured by the protein matrix. Vallee and Williams (1) coined the concept of the “entatic state” in this connection >30 years ago, and Malmström (2, 3), inspired by earlier work of Lumry (4, 5), conceived of the “protein as a rack” formalism to express similar ideas. Prime examples of preformed metal sites can be found within the class of small blue copper proteins, also known as cupredoxins. Cupredoxins function as electron carriers in biological electron transfer chains and contain a single Cu ion in their active site that shuttles, during turnover, between the Cu(I) and Cu(II) states.
At the time of their discovery, the special properties of these so-called type-1 (T1) sites were so unusual that it took coordination chemists almost a decade to realize that the specific coordination imposed by the protein on the metal is responsible for the (then) unprecedented spectroscopic and mechanistic properties of the protein (1, 6). Since then, it firmly has been established that, in T1 Cu sites, the metal is coordinated by three strong ligands (two histidines, one cysteine) arranged in a nearly trigonal planar configuration (7, 8). A fourth weak ligand (usually a methionine) occupies an axial position, whereas on the opposite side, a backbone carbonyl oxygen atom is found sometimes (as in the azurins) whose interaction with the Cu is mainly Coulombic in nature. The site appears perfectly adapted to its function [i.e., shuttling between the Cu(I) and Cu(II) states] because its geometry is a compromise between the optimal geometries of the Cu(I) and the Cu(II) states. In fact, the crystal structures of several cupredoxins reveal that the copper site geometry is virtually unaffected by a change in oxidation state (see ref. 9). The T1 coordination geometry stands in stark contrast to the square planar configuration of the T2 site, which is encountered in a variety of redox enzymes and whose spectroscopic properties are much more reminiscent of the classical coordination chemistry of Cu.
In the past 5 years, many groups have probed the uniqueness of the T1 site by replacing one or more ligands of the Cu with other amino acids by means of site-directed mutagenesis. In summary, the newly engineered sites roughly can be divided into three broad classes: (i)T1 sites with a trigonal arrangement of the three strong ligands (10, 11) and (ii)T2 sites with a square planar arrangement of four strong ligands (10–12). The new and intriguing T1.5 site (iii) with intermediate spectral properties is generated by the addition of a fourth strong ligand in the place of the weak axial group. On the basis of circumstantial evidence, the T1.5 site was correlated with a tetrahedral coordination geometry (13). The outspoken spectral features of the T1.5 site make it an excellent and important test case for any theory that claims to provide an all-encompassing description of a Cu site. To this end, structural information is needed, but, despite intensive efforts, direct structural information on a T1.5 site remained elusive until now.
The present report deals with a variant of azurin from Alcaligenes denitrificans in which the axial methionine of the Cu (Met121) has been replaced by a histidine (Met121His) (14–18). The structural characteristics of this variant have been determined in detail and are reported below. The results are remarkable for two reasons. First, they show that the protein does not constrain the metal site to the extent previously thought. Second, they represent the first successful structure determination of a T1.5 site. The crystallization of the protein was not a trivial effort in this case because the coordination properties of His121 appeared strongly pH-dependent. Thus, the structure of the Cu site varies remarkably with pH. In solutions with pH >6.5, the metal center in the M121H azurin variant exhibits all of the spectral characteristics of a protein with a T1.5 site, ascribed to the His121 being deprotonated and acting as a fourth ligand to the copper. In solutions with pH <6, the protein starts to convert into a T1 site, ascribed to His121 protonation. Because crystal growing at low pH was unsuccessful, crystals at this pH were obtained by soaking high pH crystals in low pH buffer. This soaking required special care because the low pH crystals appeared unstable and had to be flash frozen immediately on conversion of the T1.5 into the T1 site. The crystallographic results show that, in the T1.5 site, the ligands indeed are arranged in a tetrahedral manner. At a low pH, however, the site is restructured completely because of a sizable rearrangement of the loop that carries the His117 and the Met121 metal ligands. The results necessitate a revision of the entatic state concept or the protein rack mechanism.
MATERIALS AND METHODS
Crystallization, Data Collection, and Structure Determination.
Purified Met121His A. denitrificans azurin protein material was crystallized by hanging drop vapor diffusion at 21°C. Drops of 5 μl of protein solution (7 mg protein/ml) were mixed with 5 μl of buffer solution (4.0 M ammonium sulfate/1.0 M LiNO3/0.2 M acetate, pH 6.5) and equilibrated against a reservoir solution containing 2.5 M ammonium sulfate, 0.7 M LiNO3, and 0.14 M acetate (pH 6.5). After 2 days, the reservoir was adjusted to a 2.7-M ammonium sulfate concentration by adding the appropriate amount of buffer solution. Two crystal forms with different space groups and molecule content per asymmetric unit grew under the same conditions (space group C2221 for HP1 in Table 1 and P21212). Crystals for the HP2 form in Table 1 were obtained by equilibrating drops of 5 μl of protein (20 mg protein/ml) plus 5 μl of buffer (4.1 M ammonium sulfate/0.2 M sodium–potassium–phosphate, pH 6.5) against a 65% ammonium sulfate reservoir solution. The preparation of the low pH form in the crystal (LP in Table 1) was difficult to achieve and only possible starting from the P21212 pH 6.5 crystal form. As the blue low pH form is stable in the crystal for only a few minutes, it had to be fixed by shock freezing. Crystals were harvested from the drop by a cryo-loop and transferred into a cryo-buffer (2.6 M ammonium sulfate/ 0.65 M LiNO3/0.13 M acetate/30.4% glycerol, pH 3.5). After 2 min, they became blue and were shock-frozen immediately in an cold stream of evaporating liquid nitrogen.
Table 1.
Crystal parameters, data collection, structure determination, and model refinement statistics for room and liquid nitrogen temperature crystal forms of Met121His azurin at pH 6.5 and the liquid nitrogen temperature crystal form of Met121His azurin at pH 3.5
| Parameter | HP1* | HP2† | LP‡ |
|---|---|---|---|
| A Crystal parameters | |||
| Space group | C2221 | C2221 | P21212 |
| a, Å | 75.82 | 74.56 | 99.60 |
| b, Å | 74.42 | 73.57 | 100.01 |
| c, Å | 98.46 | 97.63 | 53.94 |
| Molecules/asymmetric unit | 2 | 2 | 4 |
| B Diffraction data | |||
| Resolution, Å | 1.89 | 1.91 | 2.45 |
| Total observations | 75,639 | 69,710 | 59,933 |
| Unique reflections | 21,362 | 21,288 | 19,093 |
| I/sigma§ | 8.0/1.6 | 5.8/1.8 | 3.7/2.0 |
| Rmerge§¶ | 0.076/0.455 | 0.091/0.395 | 0.059/0.182 |
| Completeness, %§ | 94.4/75.8 | 99.2/93.4 | 95.3/89.0 |
| Multiplicity | 3.5 | 3.3 | 3.1 |
| C Refinement | |||
| Resolution range, Å | 8.0–1.89/1.98–1.89 | 8.0–1.91/1.99–1.91 | 8.0–2.45/2.56–2.45 |
| Reflections in this range, n | 19,532/1,865 | 19,530/2,204 | 18,540/2,116 |
| F0 > 2.5σ(F0) | F0 > 2.5σ(F0) | no σ-cutoff | |
| R = Σ|F0-Fc|/Σ|F0|, % | 18.91/28.72 | 20.15/28.46 | 20.93/35.59 |
| Atoms, n | |||
| All nonhydrogen atoms | 2,177 | 2,323 | 4,398 |
| Nonhydrogen protein atoms | 1,956 | 1,956 | 3,912 |
| Water | 219 | 355 | 454 |
| Copper | 2 | 2 | 4 |
| Atoms in sulfate groups | — | 10 | 20 |
| Atoms in nitrate groups | — | — | 8 |
| Average temperature factor, Å2 | |||
| All nonhydrogen atoms | 25.29 | 18.14 | 24.09 |
| Nonhydrogen protein atoms | 23.09 | 14.04 | 21.99 |
| Water | 44.97 | 40.20 | 40.01 |
| Copper | 16.05 | 13.37 | 19.88 |
| Atoms in sulfate groups | — | 38.34 | 72.44 |
| Atoms in nitrate groups | — | — | 29.97 |
| Rms deviations of bonded Bs | 3.15 | 3.20 | 2.97 |
| Rms deviations from standard geometries | |||
| Bonds, Å | 0.011 | 0.011 | 0.011 |
| Angles, ° | 2.85 | 2.80 | 2.85 |
| Mean coordinate error, Å‖ | 0.21 | 0.21 | 0.30 |
| σ of mean coordinate error, Å‖ | 0.01 | 0.01 | 0.06 |
High pH form (pH 6.5) of A. denitrificans Met121His azurin measured at 289 K.
High pH form (pH 6.5) of A. denitrificans Met121His azurin measured at 100 K.
Low pH form (pH 3.5) of A. denitrificans Met121His azurin measured at 100 K.
13.96–1.89/1.95–1.89 Å for HP1; 27.74–1.91/1.98–1.91 Å for HP2; and 36.79–2.45/2.54–2.45 Å for LP.
Rmerge = ΣΣ|I(h)i − 〈I(h)〉|/ΣI(h)i, where I(h)i is the observed intensity in the ith source and 〈I(h)〉 is the mean intensity of reflection h over all measurements of I(h).
Determined from Luzzati plot (19).
The x-ray measurements were done on a Hendrix/Lentfer x-ray image–plate system (MAR Research, Hamburg, Germany) mounted on a rotating anode generator (Rigaku, Tokyo) operated at 5.4 kW (λ = CuKα = 1.5418 Å). For the cryo-measurements, a cryo-device (KGW, Karlsruhe, Germany), which was mounted on the image-plate system, was used. The x-ray intensities were processed with mosflm (19) and with programs from the CCP4 suite (20). Crystal parameters and data collection statistics are shown in Table 1 A and C. The structures of HP1 and HP2 were solved by difference Fourier techniques by using the model of the isomorphous wild-type A. denitrificans azurin (Brookhaven Protein Data Bank assession number 2AZA), omitting the His121 side chain from the structure factor calculation and refining the 2AZA model against the observed structure factors of HP1 and HP2, respectively. The LP structure solution was pursued through molecular replacement techniques by using the program amore (21). One monomer of the 2AZA structure was used as a search model. Structure factors in the 15.0–3.5 Å resolution range were used both in rotation and in translation function calculations. The correct solution had four independent molecules per asymmetric unit with a correlation factor of 72.2% and a crystallographic R factor (after rigid body refinement of the independent subunits) of 36.6%. The next highest solution had a correlation factor of 56.4% and an R factor of 45.4%. The model was refined by energy-restrained crystallographic refinement with xplor (22). The metal site geometries were refined without energy restraints. The refinement statistics of the current models are given in Table 1C. The models maintained strict geometry with deviations from ideal values for bond lengths and angles of 0.011 Å and below 2.9o, respectively. The structures refined to satisfying R factors below 21%.
RESULTS AND DISCUSSION
Crystal Structures of High pH Form.
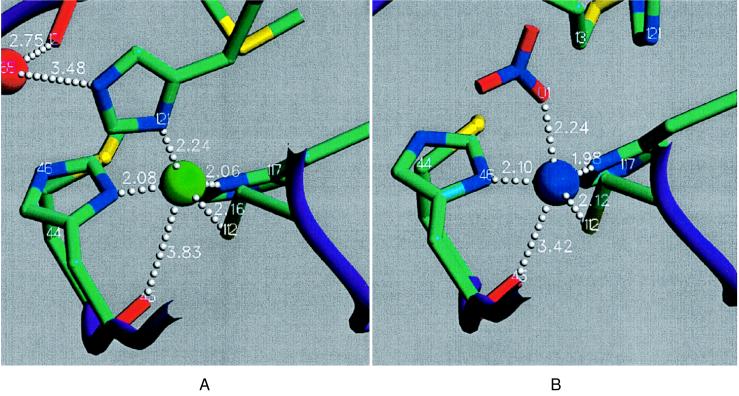
The room and liquid nitrogen temperature crystal structures of the high pH form are virtually identical and exhibit a tetrahedral copper site geometry with the deprotonated His121 as a strong equivalent ligand (Fig. 1A; Table 2), in keeping with the T1.5 classification (18). The distance of the copper from the His2Cys plane is a measure for the tetrahedrality of the copper site, and the average value for both of the high pH structures with each of the two independent molecules is 0.60 Å (σ = 0.02 Å). This average is even greater than in the T1 copper site of the copper-containing nitrite reductase from Achromobacter cycloclastes (0.54 Å) (25), which is an extreme representative of the T1-distorted tetrahedral class. In wild-type azurin, the carbonyl oxygen of Gly45 is a fifth weak ligand to the copper (2.95–3.20 Å), creating a trigonal bipyramidal coordination. The distance has increased to 3.83 Å (σ = 0.07 Å) for the two high pH structures of Met121His azurin, and 45O is, therefore, no longer coordinated to the copper. The Raman spectrum of high pH crystals shows that the same T1.5 species is present in the crystals as in solution, with the intense Cu-S stretching mode at 349 cm−1 being indicative of a significant weakening of the Cu-S(Cys) bond relative to the 408-cm−1 value for the T1 species (18). The results from this spectrum are in agreement with the increased Cu-S(Cys) bond length of 2.16 Å in the high pH structures compared with the 2.12-Å value for the low pH T1 form (Table 2).
Figure 1.
Copper site of A. denitrificans Met121His azurin. (A) High pH form. The indicated distances are the mean values over the subunits in HP1 and HP2. (B) Low pH form. The displayed distances are the mean values between subunits C and D of LP. Image produced with setor (24).
Table 2.
Copper site geometries of high pH and low pH forms of A. denitrificans Met121His azurin
| HP1
|
HP2
|
X* | σ | LP
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SUA | SUB | SUA | SUB | SUA | SUB | X† | σ | SUC | SUD | X‡ | σ | |||
| Bond distances, Å | ||||||||||||||
| Cu-112Sγ | 2.16 | 2.16 | 2.25 | 2.06 | 2.16 | 0.07 | 2.25 | 2.06 | 2.16 | 0.08 | 2.06 | 2.17 | 2.12 | 0.06 |
| Cu-46Nδ | 2.02 | 2.02 | 2.06 | 2.20 | 2.08 | 0.07 | 2.06 | 2.13 | 2.08 | 0.06 | 2.08 | 2.11 | 2.10 | 0.02 |
| Cu-117Nδ | 2.14 | 2.02 | 2.02 | 2.05 | 2.06 | 0.05 | 1.95 | 1.89 | 2.01 | 0.08 | 1.94 | 2.02 | 1.98 | 0.04 |
| Cu-121Nδ | 2.28 | 2.16 | 2.24 | 2.28 | 2.24 | 0.05 | 2.26 | 2.26 | 2.25 | 0.04 | — | — | — | — |
| Cu-45O | 3.93 | 3.84 | 3.77 | 3.77 | 3.83 | 0.07 | 3.71 | 3.76 | 3.80 | 0.07 | 3.44 | 3.39 | 3.42 | 0.02 |
| Cu-nitrate O | — | — | — | — | — | — | — | — | — | — | 2.12 | 2.35 | 2.24 | 0.12 |
| Bond angles, ° | ||||||||||||||
| 112Sγ-Cu-46Nδ | 129.0 | 128.0 | 130.2 | 131.0 | 129.6 | 1.1 | 127.3 | 127.6 | 128.9 | 1.4 | 134.6 | 134.8 | 134.7 | 0.1 |
| 112Sγ-Cu-117Nδ | 109.3 | 103.8 | 108.7 | 110.8 | 108.2 | 2.6 | 107.0 | 104.1 | 107.3 | 2.6 | 122.3 | 122.8 | 122.6 | 0.25 |
| 112Sγ-Cu-121Nδ | 100.5 | 101.8 | 106.2 | 101.6 | 102.5 | 2.2 | 108.5 | 115.1 | 105.6 | 5.1 | — | — | — | — |
| 46Nδ-Cu-117Nδ | 98.5 | 98.7 | 98.3 | 97.4 | 98.2 | 0.5 | 96.3 | 97.6 | 97.8 | 0.8 | 97.3 | 92.9 | 95.1 | 2.2 |
| 46Nδ-Cu-121Nδ | 91.9 | 94.1 | 89.3 | 87.1 | 90.6 | 2.6 | 84.8 | 81.4 | 88.1 | 4.0 | — | — | — | — |
| 117Nδ-Cu-121Nδ | 130.4 | 134.6 | 125.7 | 131.3 | 130.5 | 3.2 | 134.0 | 131.0 | 131.2 | 2.9 | — | — | — | — |
| 46Nδ-Cu-nitrate O | — | — | — | — | — | — | — | — | — | — | 77.1 | 88.0 | 82.6 | 5.5 |
| 117Nδ-Cu-nitrate O | — | — | — | — | — | — | — | — | — | — | 97.4 | 96.6 | 97.0 | 0.4 |
| 112Sγ-Cu-nitrate O | — | — | — | — | — | — | — | — | — | — | 114.7 | 111.5 | 113.1 | 1.6 |
Mean value of HP1 and HP2 subunits.
Mean value of HP1, HP2, and LP subunits A and B.
Mean value of LP subunits C and D.
Crystal Structure of Low pH Form.
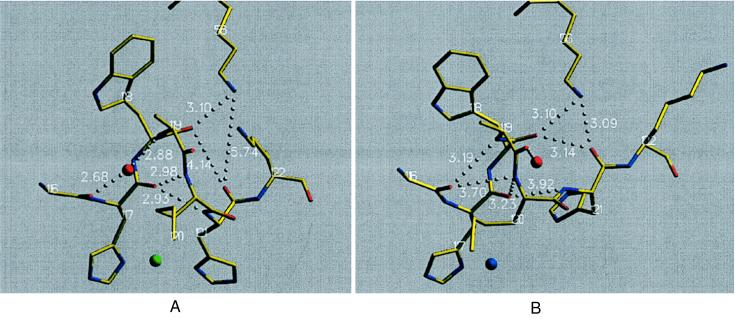
The crystal structure of the low pH form, in which four independent molecules were present, showed the copper site geometry in two molecules (A and B) identical to that in the high pH form and in the two other molecules (C and D) changed (Fig. 1B; Table 2). The protonation of His121 in subunits C and D not only causes a break of the Nδ-Cu bond but also generates a major rearrangement of the main chain between residues 120 and 123 with average rmsd values of 3.76 Å (all atoms) and 2.75 Å (main chain plus Cβ atoms) for both relevant molecules (Fig. 2). This rearrangement is a transition of the one-turn helical structure from residues 116 to 121, which is intermediate between 310- and α-helical in the high pH form (Fig. 2A), into a turn of 310-helical character in the low pH form (Fig. 2B). In the high pH form (Fig. 2A), 116O does not form a direct hydrogen bond to possible amide nitrogens in positions i+3 and i+4, but a water molecule bridges 116O with 119N. 117O is hydrogen-bonded to the amide nitrogens of residues 120 and 121, creating the intermediate 310-α-helical structure of this turn. This arrangement enables the His121 side chain to coordinate to the copper. Nζ of Lys56 is hydrogen-bonded to 118O, and the 121O has distances of 4.14 Å to 118O and 5.24 Å to 56Nζ. In the low pH form (Fig. 2B), the turn adopts a 310-helix conformation with hydrogen bond is from 116O to 119N (3.19 Å) and 117O to 120N (3.23 Å). The corresponding i+4-hydrogen bonds are no longer present (3.70 Å and 3.92 Å, respectively). His121 and Lys122 have been moved in such a way that Nδ of His121 is ≈5.27 Å apart from the copper ion and 121O is now hydrogen-bonded to Lys56 at a distance of 3.14 Å from 118O. This movement is accommodated by Gly123 (Fig. 3), which acts as a hinge element in this process. It is well known that glycine residues are pivotal residues in flexible protein structural elements (26). At Thr124, the C-terminal β-strand remains unchanged in its position and conformation in both pH forms. Additionally, Fig. 3 displays the final omit difference electron density for the polypeptide stretch 120–123 in subunit D.
Figure 2.
Polypeptide stretch of A. denitrificans Met121His azurin that undergoes the conformational change on protonation of His121 at low pH (3.5). (A) High pH form. The indicated distances are mean values between subunits A and B of both HP and LP. (B) Low pH form. The displayed distances are the mean values between subunits C and D of LP. Image produced with setor (24).
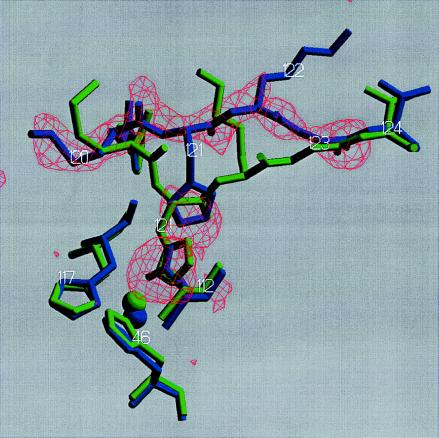
Figure 3.
Overlay of the low pH form (subunit D of LP, blue) onto high pH form (subunit A of LP, green) of the copper sites and the polypeptide stretch 120–124. The included 2.45-Å resolution Fo–Fc omit electron density map was calculated with residues 120–123, and nitrate in subunits D was removed and running a 200-cycle positional refinement before map calculation. The map has been contoured at 3.0 σ. Image produced with setor (24).
Different crystal packing contacts of the individual subunits in the P21212 crystal form seem to determine the different copper coordinations in the low pH P21212 crystal form (LP) with the spectroscopic high pH form in subunits A and B and the spectroscopic low pH form in subunits C and D of LP. In the P21212 crystals, the subunits are arranged as two dimers (subunits A and C as well as B and D). The monomers of each dimer are linked via a local twofold axis, and the dimers are separated by an approximate C centering. To simulate the situation in the high pH P21212 crystal form (the starting point for generating the low pH crystal form), subunits A and B of LP were superimposed onto subunits C and D of LP, respectively, with lsqman (27). A crystal contact calculation of this simulated high pH P21212 crystal form showed that Lys122, which is one of the residues that is most shifted as a result of the transition, has different crystal contacts in the various subunits. In subunits A and B of the simulated structure, Lys122 is in van der Waals contact with Lys38 of a neighboring molecule, which may prevent the movement of the polypeptide chain, thus also preventing protonation of His121. On the other hand, Lys122 of subunits C and D of the simulated form have Lys38 of a neighboring molecule as the closest contact partner at a distance of >4.5 Å. In this case, the movement of the polypeptide chain, as observed in subunits C and D of LP, may occur, and His121 can be protonated. The rearrangement of the loop seen in the crystal can be observed in solution at high pH as a time-dependent phenomenon that gives rise to exchange broadening in the total correlation spectroscopy spectra of the solution and to the occurrence of cross peaks in the exchange spectroscopy spectra (14, 18).
The copper coordination of the (blue) spectroscopic low pH form (T1), as realized in subunits C and D of LP, is axially distorted tetrahedral with an axial oxygen ligand originating from a coordinating nitrate molecule (Fig. 1B and Table 2). The average distance of the copper ion from the His2Cys plane for subunits C and D is 0.32 Å. The out-of-plane movement is in the direction of the coordinating nitrate oxygen. Furthermore, the mean 45O–Cu distance is 3.42 Å, which means that the Gly45 carbonyl oxygen does not play a significant role in the coordination of the Cu. The coordination geometry is characteristic of T1 sites with an oxygen donor in the axial position as determined from the x-ray structures of oxidized M121Q-azurin (28) and cucumber stellacyanin (29). The axial position of the nitrate molecule is substituted by a water or hydroxyl oxygen in the corresponding nitrate-free buffer, as initially suggested on the basis of spectroscopic evidence (18) and subsequently confirmed by nuclear magnetic relaxation–dispersion and perturbed angular correlations of γ-rays experiments (15, 16).
Final Remarks.
Two important conclusions can be drawn from the results presented above. The first one is about the correlation of spectroscopic features with site structure. In the recent past, several attempts have been made to use the spectroscopic fingerprint of a Cu site as a clue to its structure. The basis for this was the initial observation that the T1 and the (cysteine-containing) T2 sites have markedly different spectroscopic features (13) as observed in the optical spectra (main absorption band near 600 vs. 400 nm), in the resonance Raman spectra (main Cu-S frequencies near 400 vs. 300 cm−1), and in the low-field region of the electron paramagnetic resonance spectra (A∥ values near 60 vs. 150 Gauss), showing that the two sites correspond to clearly different coordination geometries. In the course of time, the scheme tentatively was extended to correlate small variations in spectroscopic parameters with subtle variations in structure, but it could never be put to a rigorous test until now. An important criterion for the predictive power of the scheme was conceived when a site was observed (T1.5) with a spectroscopic fingerprint that was intermediate between the T1 and T2 fingerprints (two medium strength optical absorption bands near 420 and 560 nm; resonance Raman frequencies in the intermediate range near 350 cm−1; and electron paramagnetic resonance A∥ values intermediate near 100 Gauss). The site geometry was predicted to be close to tetrahedral. The present results bear out this prediction and, thus, in a decisive way vindicate the use of spectroscopic features for the structural characterization of single Cu sites in proteins.
Second, the present results shed new light on the role of the protein in determining the structure of a metal site. The unusual and unexpected (from a coordination chemist’s point of view) coordination geometry of the T1 Cu site led a long time ago to the idea that the protein matrix fixes the scaffold in which the metal subsequently is accommodated. The observations described above necessitate an important adjustment of this concept. What we see is that, when the attractive interaction between the metal (Cu in our case) and one of its ligands (His121) is abolished or possibly even reversed (protonation of His121), the protein matrix responds by a major local rearrangement of the protein structure and the introduction of external ligands (nitrate, hydroxyl, water). The structural differences between the high- and low-pH forms of M121H azurin can be accounted for by protein flexibility in positioning the axial ligand and by the facile movement of the Cu along the axial coordinate. The conclusion must be that the metal-binding energy can be a decisive factor in the formation of a structurally stable metal site and that the metal, therefore, codetermines the structure of its own site.
ABBREVIATIONS
- T
type
- HP1
high pH form (pH 6.5) of A. denitrificans Met121His azurin measured at 289K
- HP2
high pH form (pH 6.5) of A. denitrificans Met121His azurin measured at 100 K
- LP
low pH form (pH 3.5) of A. denitrificans Met121His azurin measured at 100 K
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference nos. 1A4A, 1A4B, and 1A4C).
References
- 1.Vallee B L, Williams R J P. Proc Natl Acad Sc USA. 1968;59:498–505. doi: 10.1073/pnas.59.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindskog S, Malmström B G. J Biol Chem. 1962;237:1129–1138. [PubMed] [Google Scholar]
- 3.Malmström B G, Vänngard T. J Mol Biol. 1960;2:118–124. doi: 10.1016/s0022-2836(62)80074-6. [DOI] [PubMed] [Google Scholar]
- 4.Lumry R, Eyring H. J Phys Chem. 1954;58:110–120. [Google Scholar]
- 5.Eyring H, Lumry R, Spikes J D. In: Mechanism of Enzyme Action. McElroy W D, Glass B, editors. Baltimore: Johns Hopkins Univ. Press; 1956. pp. 123–140. [Google Scholar]
- 6.Colman P M, Freeman H C, Guss J M, Murata M, Norris V A, Ramshaw J A, Venkatappa M P. Nature (London) 1978;272:319–324. [Google Scholar]
- 7.Sykes A G. Adv Inorg Chem. 1991;36:377–408. [Google Scholar]
- 8.Adman E T. Adv Protein Chem. 1991;42:145–198. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 9.Messerschmidt A. Struct Bonding (Berlin) 1998;90:38–68. [Google Scholar]
- 10.den Blaauwen T, Hoitink C W G, Canters G W, Han J, Loehr T M, Sanders-Loehr J. Biochemistry. 1993;32:12455–12464. doi: 10.1021/bi00097a025. [DOI] [PubMed] [Google Scholar]
- 11.Andrew C R, Yeom H, Valentine J S, Karlsson B G, Bonander N, van Pouderoyen G, Canters G W, Loehr T M, Sanders–Loehr J. J Am Chem Soc. 1994;116:11489–11498. [Google Scholar]
- 12.Faham S, Mizoguchi T J, Adman E T, Gray H B, Richards J H, Rees D C. J Biol Inorg Chem. 1997;2:464–469. [Google Scholar]
- 13.Canters G W, Gilardi G. FEBS Lett. 1993;325:39–48. doi: 10.1016/0014-5793(93)81410-2. [DOI] [PubMed] [Google Scholar]
- 14.Kroes S J, Salgado J, Parigi G, Lichinat C, Canters G W. J Biol Inorg Chem. 1996;1:551. [Google Scholar]
- 15.Salgado J, Kroes S J, Berg A, Moratal J M, Canters G W. J Biol Chem. 1998;273:177–185. doi: 10.1074/jbc.273.1.177. [DOI] [PubMed] [Google Scholar]
- 16.Danielsen E, Kroes S J, Canters G W, Bauer R, Hemmingsen L, Singh K, Messerschmidt A. Eur J Biochem. 1997;250:249–259. doi: 10.1111/j.1432-1033.1997.0249a.x. [DOI] [PubMed] [Google Scholar]
- 17.Salgado J, Jiminez H R, Moratal J M, Kroes S J, Warmerdam G C M, Canters G W. Biochemistry. 1996;35:1810–1819. doi: 10.1021/bi951748a. [DOI] [PubMed] [Google Scholar]
- 18.Kroes S J, Hoitink C W G, Andrew C R, Ai J, Sanders-Loehr J, Messerschmidt A, Hagen W R, Canters G W. Eur J Biochem. 1996;240:342–351. doi: 10.1111/j.1432-1033.1996.0342h.x. [DOI] [PubMed] [Google Scholar]
- 19.Leslie A G W. In: Crystallographic Computing. Moras D, Podjarny A D, Thierry J C, editors. Oxford: Oxford Univ. Press; 1990. pp. 50–61. [Google Scholar]
- 20.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 21.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 22.Brünger A T. xplor, Version 3.1, A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 23.Luzzati V. Acta Crystallogr A. 1952;5:802–810. [Google Scholar]
- 24.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 25.Adman E T, Godden J W, Turley S. J Biol Chem. 1995;270:27458–27474. doi: 10.1074/jbc.270.46.27458. [DOI] [PubMed] [Google Scholar]
- 26.Hammann C, van Pouderoyen G, Nar H, Gomis Rüth F-X, Messerschmidt A, Huber R, den Blaauwen T, Canters G W. J Mol Biol. 1997;266:357–366. doi: 10.1006/jmbi.1996.0764. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt, G. J. & Jones, T. A. (1994) ESF/CCP 4 Newsletter, 9–14.
- 28.Romero A, Hoitink C W G, Nar H, Huber R, Messerschmidt A, Canters G W. J Mol Biol. 1993;229:1007–1021. doi: 10.1006/jmbi.1993.1101. [DOI] [PubMed] [Google Scholar]
- 29.Hart P J, Nerssisian A M, Herrmann R G, Nalbandyan R M, Valentine J S, Eisenberg D. Protein Sci. 1996;5:2175–2183. doi: 10.1002/pro.5560051104. [DOI] [PMC free article] [PubMed] [Google Scholar]