Innate immunity is an ancient form of host defense that is shared by almost all multicellular organisms (1, 2). However, it is not a redundant defense mechanism, and recent evidence has shown that innate immunity not only provides a first line of antimicrobial host defense, but also has a profound impact on the establishment of adaptive immune responses (1, 3). Upon infection, microorganisms are first recognized by cells of the host innate immune system, such as phagocytic leukocytes, endothelial and mucosal epithelial cells, and professional antigen-presenting cells. Recognition of pathogens is primarily mediated by a set of germline-encoded molecules on innate immune cells that are referred to as pattern recognition receptors (PRRs) (3). Well characterized PRRs include CD14, β2-integrins (CD11/CD18), C-type lectins, macrophage scavenger receptors, and complement receptors (CR1/CD35, CR2/CD21) (3). These PRRs are expressed as either membrane-bound or soluble proteins that recognize invariant molecular structures called pathogen-associated molecular patterns (PAMPs) that are shared by many pathogens but not expressed by hosts (3). Examples of PAMPs include LPS, bacterial lipoprotein (BLP), peptidoglycan (PGN) lipoteichoic acid (LTA), unmethylated CpG DNA of bacteria, lipoarabinomannan (LAM) of mycobacteria, and mannans of yeast (3).
Recognition of PAMPs by PRRs results in the activation of different intracellular signaling cascades that in turn lead to the expression of various effector molecules (3). One group of effector molecules consists of reactive oxygen and nitrogen intermediates and various antimicrobial peptides that have direct microbicidal activity and collectively provide immediate protection for hosts. Another group includes cytokines, chemokines, adhesion molecules, and acute phase proteins that are involved in inflammation and early host defense as well as the development of adaptive immune responses. The third group consists of the costimulatory molecules B7.1 and B7.2, which bind CD28 on T cells and act as the second signal for T-cell activation. Therefore, signaling by the PRRs helps to bridge innate and adaptive immunity and allows the host to cope more efficiently with microbial infection. In keeping with the important role that innate immunity plays in protecting multicellular organisms from infection, components of the innate immune response, including pathogen recognition molecules, signal transduction pathways, and downstream effector molecules, are all evolutionarily conserved and are used by insects, plants, and mammals (2).
Recent studies on the recognition of microbial PAMPs have highlighted the critical role of one group of PRRs, the Toll-like receptors (TLRs), in pathogen recognition and host defense. These TLRs are distinguished from other PRRs by their ability to recognize and, more significantly, discriminate between, different classes of pathogens (reviewed in refs. 4, 5). Engagement of TLRs by pathogens leads to the activation of innate immune responses (5), and a major signaling target of the TLRs is activation of the transcription factor NF-κB, a key regulator of immune and inflammatory responses (reviewed in refs. 6–8). Interestingly, TLR-mediated NF-κB activation is also an evolutionarily conserved event that occurs in phylogenetically distinct species ranging from insects to mammals (5, 9, 10). Here, we focus on the role of the conserved TLR/NF-κB signaling pathway in innate immunity, as well as its impact on adaptive immune responses.
NF-κB/Rel proteins: key regulators of innate immune responses
NF-κB was originally identified as a transcription factor that bound to an enhancer element in the gene for the Igκ light chain and was believed to be B cell–specific (reviewed in refs. 6–8). However, subsequent studies revealed that it is ubiquitously expressed and plays a central role in regulating the expression of many genes involved in immune, inflammatory, and apoptotic processes (6–8). NF-κB can be activated by different stimuli such as microbial products, proinflammatory cytokines, T- and B-cell mitogens, and physical and chemical stresses (6–8). NF-κB in turn regulates the inducible expression of many cytokines, chemokines, adhesion molecules, acute phase proteins, and antimicrobial peptides (6–8).
NF-κB represents a group of structurally related and evolutionarily conserved proteins. So far, five mammalian NF-κB proteins named Rel (c-Rel), RelA (p65), RelB, NF-κB1 (p50 and its precursor p105), and NF-κB2 (p52 and its precursor p100) have been described (6, 8). NF-κB/Rel proteins can exist as homo- or heterodimers, and although most NF-κB dimers are activators of transcription, the p50/p50 and p52/p52 homodimers repress the transcription of their target genes (6, 8). In Drosophila, three NF-κB homologs named Dorsal, Dif, and Relish have been identified and characterized (5, 6). Structurally, all NF-κB/Rel proteins share a highly conserved NH2-terminal Rel homology domain (RHD) that is responsible for DNA binding, dimerization, and association with inhibitory proteins known as IκBs (6, 8). In resting cells, NF-κB/Rel dimers are bound to IκBs and retained in an inactive form in the cytoplasm. Like NF-κB, IκBs are also members of a multigene family containing seven known mammalian members including IκBα, IκBβ, IκBγ, IκBε, Bcl-3, the precursor Rel-proteins, p100, and p105, and one Drosophila IκB named Cactus (6, 8). The IκB family is characterized by the presence of multiple copies of ankyrin repeats, which are protein-protein interaction motifs that interact with NF-κB via the RHD. Upon appropriate stimulation, IκB is phosphorylated by IκB kinases (IKKs), polyubiquitinated by a ubiquitin ligase complex, and degraded by the 26S proteosome (6, 8). Consequently, NF-κB is released and translocates into nucleus to initiate gene expression.
NF-κB regulates the expression of a wide variety of genes that play critical roles in innate immune responses (6, 7). Such NF-κB target genes include those encoding cytokines (e.g., IL-1, IL-2, IL-6, IL-12, TNF-α, LTα, LTβ, and GM-CSF), adhesion molecules (e.g., ICAM, VCAM, endothelial leukocyte adhesion molecule [ELAM]), acute phase proteins (e.g., SAA), and inducible enzymes (e.g., iNOS and COX-2) (6–8). In addition, it has been demonstrated recently that several evolutionarily conserved antimicrobial peptides, e.g., β-defensins, are also regulated by NF-κB (11), a situation similar to Drosophila (2, 5). Besides regulating the expression of molecules involved in innate immunity, NF-κB also plays a role in the expression of molecules important for adaptive immunity, such as MHC proteins, and the expression of critical cytokines such as IL-2, IL-12 and IFN-γ (6). Finally NF-κB plays an important role in the overall immune response by affecting the expression of genes that are critical for regulating the apoptotic process, such as c-IAP-1 and c-IAP-2, Fas ligand, c-myc, p53, and cyclin D1 (12).
TLRs, a family of major pathogen recognition molecules
TLRs represent a growing family of transmembrane proteins characterized by multiple copies of leucine-rich repeats (LRRs) in the extracellular domain and a cytoplasmic Toll/IL-1R (TIR) motif. As their name suggests, TIR motifs of TLRs exhibit significant homology to the intracellular signaling domain of the type I IL-1 receptor (IL-1RI) (Figure 1) and therefore, TLRs are thought to belong to the IL-1R superfamily (10). Although the IL-1R and TLRs differ in their extracellular domains, the presence of the TIR domain allows both receptors to activate similar intracellular signaling pathways (see below).
Figure 1.
Sequence alignment of the intracellular signaling domains of representative members of TLR/IL-1 receptor family. Conserved residues are boxed, with identical amino acids shaded in dark gray and similar amino acids shaded in light gray. The COOH-terminal region that is indispensable for TLR- and IL-1–mediated signaling is underlined. The proline residue that is conserved among eight of the nine human TLRs, which is critical for signaling induced by TLR2 and TLR4, is marked with a triangle.
TLRs are evolutionarily conserved and their congeners have been found in insects, plants, and mammals. Drosophila Toll (dToll) was the first member of the TLR family to be identified, and was initially characterized as a developmental protein governing the formation of the dorsal-ventral axis in Drosophila (9). However, subsequent studies revealed that dToll also plays a key role in triggering innate immune responses against fungal infection in adult flies (5, 9). To date, more than ten distinct Toll-like sequences, homologous to the highly conserved cytoplasmic domain sequence of dToll, have been identified in the largely completed Drosophila genomic sequence (http://www.fruitfly.org/blast). Among the ten potential Toll-like proteins, only dToll and 18-Wheeler have been studied with regard to their role in resistance to infection (5). In humans, nine full-length TLR sequences have also been deposited in GenBank, while six other members remain partially characterized (5, 10). More members are expected to appear upon completion of the human genome project. In addition, plants express Toll-like molecules, including N, L6, RPP1, and RPP5, that are involved in disease resistance (13). Surprisingly, these plant homologs are exclusively cytoplasmic proteins, although they share homologous LRR and TIR domains with their relatives in insects and mammals (13).
Consistent with their role in pathogen recognition and host defense, mammalian TLRs are strategically expressed in monocytes/macrophages, neutrophils, dendritic cells, intestinal epithelial cells and endothelial cells — cell types that are immediately accessible to microorganisms upon infection (14). TLR2 and TLR4 have also been found in B and T cells, a possible indication of their role in modulating adaptive immune responses (14).
CD14: a coreceptor for recognition of microbial products
It has been known for almost a decade that host recognition of microbial products such as LPSs, PGNs, BLPs, LAM, and LTA is principally mediated by either a membrane-bound or soluble form of the glycoprotein CD14 (reviewed in refs. 15, 16). A serum factor named LPS-binding protein (LBP) is believed to act as a shuttle to transfer microbial products to CD14 (15, 16). Binding of microbial products to CD14 triggers production of various proinflammatory cytokines including IL-1 and TNF-α. Because membrane-bound CD14 is a glycosyl phosphatidyl inositol–anchored protein devoid of an intracellular domain, the mechanism by which its signals are transduced has remained a mystery. However, several independent lines of evidence supported the hypothesis that LPS functions by interacting with a second signaling receptor (reviewed in ref. 16). First, whereas both LPS and LAM can activate CD14-transfected mouse B cells, LAM alone failed to activate CD14-transfected hamster fibroblasts, implying that hamster, but not mouse, LPS-unresponsive cells lack a specific signal transducer for LAM. Second, enzymatically deacylated LPS or the synthetic tetra-acyl lipid A analog (lipid IVA), activates LPS-responsive mouse and hamster cells but fails to do so in human cells, regardless of the species of CD14 expressed on the cell surface (16). These studies suggested that species-specific recognition of LPS is not mediated by CD14 but by another, as yet unidentified molecule(s).
Perhaps the most compelling evidence for the existence of CD14-associated signal transducers came from genetic studies. In CD14-deficient mice, LPS can activate monocytes to produce TNF-α and IL-6, albeit at concentrations two to three times higher than those required for wild-type mice (reviewed in ref. 16). In addition, both C3H/HeJ and C57BL/10ScCr inbred mice are highly tolerant to LPS challenge (17), and even with normal levels of CD14 expressed on the cell surface, macrophages from these mice failed to respond to LPS. The ability of these mice to respond to LPS had been genetically linked to the Lps locus on chromosome 4 (17). Intriguingly, these mice respond normally to many other microbial products, such as LAM. Taken together, these observations clearly indicated that LPS responsiveness was not solely dependent upon CD14, but that other cell surface molecules help to mediate LPS signaling. Moreover, LPS and other microbial products most likely employ different receptors to transduce their signals, although CD14 might be a common shared element of the different receptors.
TLRs act in concert with CD14 to confer responsiveness to microbial products
Studies of mammalian TLRs have now provided tantalizing evidence that TLRs are the CD14-associated signal transducers for different classes of microbial products. In addition, TLRs appear to help innate immune cells recognize and distinguish between pathogens, and initiate signaling cascades for mounting appropriate responses to invading pathogens. TLRs also help bridge innate and adaptive immunity by inducing various costimulatory and effector molecules.
Among the nine known mammalian TLRs, only TLR2 and TLR4 have been extensively characterized. The biological functions of other members remain largely unknown, since TLR1, -3, -5, and -6 fail to impart signals from microbial products when expressed in unresponsive cells (4, 5, 17). The first identified human TLR, now called TLR4, activates NF-κB and induces expression of IL-1, IL-6, IL-8, and the costimulatory molecule B7 (18). Later, TLR2 and TLR4 were found to activate NF-κB in response to LPS stimulation, a response that is enhanced by the presence of CD14 and LBP (4, 19, 20). TLR2 forms homodimers and physically associates with CD14 (21). It triggers innate host defense by inducing the production of reactive oxygen species (22) and the transcription of IL-12 and nitric oxide synthase (23) in response to lipoproteins of Mycobacterium tuberculosis. More convincingly, the gene that was previously known as Lps in C3H/HeJ and C57BL/10ScCr mice has recently been determined to be Tlr4 (reviewed in ref. 17). C3H/HeJ mice have a point mutation in the coding region of the Tlr4 gene, resulting in a nonconservative substitution of proline with histidine at position 712, whereas C57BL/10ScCr mice are homozygous for a null mutation. Indeed, proline 712 is conserved among nine known mammalian TLRs with the exception of TLR3 (Figure 1). Targeted disruption of the Tlr4 gene renders normal mice hyporesponsive to LPS, a phenotype similar to LPS-tolerant mice (24). Furthermore, mutations cosegregating in LPS-hyporesponsive humans were mapped to positions Asp299Gly and Thr399Ile in TLR4 and lie within the extracellular domain of TLR4 (25). These lines of genetic evidence strongly suggest that TLR4 is the primary signal transducer for LPS. On the other hand, TLR2 is either a low affinity receptor for LPS (19, 20), or is not a receptor for LPS, but instead responds to low levels of contaminating BLPs in commercial LPS preparations (26).
Differential recognition of pathogens by TLRs
Why are there so many TLRs? One possibility is that different TLRs recognize different classes of PAMPs that characterize different pathogens. Precedent for such a scenario exists in Drosophila, where Toll-like proteins are differentially activated in response to infection. For example, Toll principally mediates antifungal responses, whereas 18-Wheeler is directed against bacterial pathogens (reviewed in ref. 5). Interestingly, Drosophila also responds differently to various classes of pathogens by producing distinct classes of antimicrobial peptides (5). For instance, drosomycin, a peptide with fungicidal activity, is rapidly induced in response to a fungal infection, but is not induced during Gram-positive or -negative infections (5). Drosophila NF-κB homologs, Dorsal, Dif, and Relish, also appear to be differentially activated in response to challenges with different classes of pathogens (5). It remains to be determined whether infection with different classes of bacteria, or engagement of distinct mammalian TLRs, results in the activation of separate combinations of NF-κB/Rel dimers, thereby eliciting a distinct profile of effector molecules and leading to defense responses appropriate for specific kinds of invading bacteria.
This property of differential recognition of pathogens also appears to be conserved in mammalian systems. Biochemical studies and investigations using mice deficient in TLR2 and TLR4 indicated that TLR4 is used primarily for mediating cellular signaling induced by LPS and LTA. In contrast, TLR2 mediates responses to PGNs and cell wall components of Gram-positive bacteria, mannans of yeasts, LAM of mycobacteria, BLPs of M. tuberculosis, mycoplasma, and the Lyme disease–causing spirochete Borrelia burgdorferi (4, 17, 27). In addition, species-specific recognition of lipid IVA also illustrates the principle of differential recognition of bacterial products. Thus, human TLR4 does not respond to lipid IVA, whereas both mouse and hamster TLR4 respond to lipid IVA, leading to the production of proinflammatory cytokines (28, 29).
The ability of TLR4 to recognize and discriminate between the fine structural differences within the lipid A portion of LPS supports the notion that TLR4 physically interacts with lipid A (28, 29). Upon binding to CD14, LPS might be either transferred to TLR4, or form a ternary molecular complex with TLR4 and CD14. However, such direct interaction with microbial products is in marked contrast to the situation in Drosophila, where the ligand for dToll is Spatzle, an endogenous protein generated from a proteolytic cascade initiated during development or upon bacterial infection (5, 9). Hence, the actual receptor for bacterial products is upstream of Toll in the signaling pathway. Given the conservation of the Toll signaling system in insects and mammals, a mammalian homolog of Spatzle has been predicted as the ligand for TLRs instead of bacterial products. However, no such mammalian protein has been identified to date (17). Intriguingly, heat shock protein 60 (HSP60) (30) and HSP70 (31) were recently found to use CD14 and TLR4 for transducing signals leading to generation of proinflammatory mediators, suggesting that the HSPs might represent a novel class of putative endogenous ligands for TLRs.
TLR-mediated signaling pathways
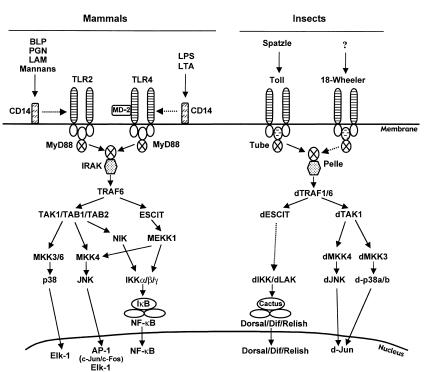
Upon engagement of TLRs with microbial products, several intracellular signal transduction pathways are activated and initiated. Among the most prominent and best characterized of these is that leading to NF-κB activation. TLR-induced NF-κB activation represents a critical component of an ancient innate host defense system, which is phylogenetically conserved in insects and mammals (5, 9, 10) (Figure 2). Upon activation, TLRs most likely form homodimers, resulting in a conformational change in the cytoplasmic TIR domain and subsequent recruitment of an adapter named MyD88 (5, 10). MyD88 associates with the TLR via a homophilic interaction using the TIR domains. The death domain of MyD88 then recruits downstream IL-1 receptor–associated kinase (IRAK) to the receptor complex (10). IRAK is then autophosphorylated and dissociated from the receptor complex and recruits TNF receptor–associated factor 6 (TRAF6) that in turn activates downstream kinases. Several such kinases have been found to be involved in TLR/NF-κB signaling pathways including NF-κB–inducing kinase (NIK) and mitogen-activated protein kinase/ERK kinase kinase 1 (MEKK1) (8).
Figure 2.
TLR-mediated signaling pathways leading to activation of NF-κB and MAPK. TLR2 is activated in response to BLPs, PGNs of Gram-positive bacteria, LAM of mycobacteria, and mannans of yeasts. TLR4 is activated by LPS of Gram-negative bacteria and LTA of Gram-negative bacteria. A secreted small molecule MD-2 is essential for TLR4 signaling. Whether bacterial products directly bind to TLRs, or whether they bind through CD14, remains to be fully established. In Drosophila, Toll is activated by Spatzle, an endogenous protein generated by a protease cascade during development and fungal infection. A different Toll-like molecule, 18-Wheeler, is activated upon bacterial infection. Whether Spatzle is also a ligand for 18-Wheeler remains to be determined. It is important to note that NF-κB and MAPK can also be activated independently of MyD88, and TLR2 can lead to apoptosis. However, the details of the pathways involved remain to be characterized.
The mechanism by which NIK or MEKK1 is activated is not well understood. Recently, TRAF6 was shown to interact with another protein, termed evolutionarily conserved signaling intermediate in Toll pathway (ECSIT), which aids in processing MEKK1 to the active form (32). NIK, on the other hand, can be phosphorylated and activated by TGF-β–activated kinase 1 (TAK1), which binds to TRAF6 through the binding proteins TAB1 and TAB2 (33, 34). Activated MEKK1 or NIK are individually capable of activating the IKK complex (8). Subsequently, IκB is phosphorylated and degraded, leading to nuclear translocation of NF-κB and initiation of gene transcription. It is not known whether all TLRs use the same signaling intermediates to activate NF-κB, but Shimazu et al. (35) have identified one subtle difference between TLR2- and TLR4-mediated signaling, namely, the requirement for the secreted molecule MD-2 to confer LPS-responsiveness on TLR4 but not TLR2 (19, 20, 22, 23).
Interestingly, insects and plants have also evolved a homologous TLR/NF-κB signaling system that is employed for host defense (Figure 2). In Drosophila, Toll is activated upon binding of Spatzle, as discussed above (9). An adapter protein, Tube, and a serine/threonine kinase homologous to IRAK named Pelle, is then recruited to the receptor (5, 9). Similar to the mammalian system, a Drosophila TRAF protein interacts with Pelle (36, 37), presumably followed by its association with Drosophila ECSIT (32). This activation of these signaling pathways ultimately leads to the phosphorylation and degradation of Cactus, leading to nuclear translocation of Dorsal, Dif, and Relish. Recently, a Drosophila IKK, termed LPS-activated kinase (LAK), was cloned and found to phosphorylate Cactus (38).
In addition to NF-κB activation, TLRs can also initiate mitogen-activated protein kinase (MAPK) signaling cascades and thus activate multiple transcription factors, including activator protein 1 (AP-1) and Elk-1 (5, 17, 39) (Figure 2). It is well established that LPS stimulation of monocytes/macrophages induces phosphorylation of p38, ERK1/2, and c-Jun NH2-terminal kinase (JNK) (15), although the molecular mechanisms by which these MAPKs are activated remain largely unknown. However, several molecules, including TAK1/TAB1, have been identified that are capable of activating the MAPKKs MKK3/6 and MKK4, which, in turn, activate p38 and JNK, respectively (8, 33). MEKK1 also activates the JNK pathway by phosphorylating MKK4. However, as demonstrated in MyD88-deficient mice, MAPKs can be activated independent of MyD88 (40). Drosophila has also evolved a homologous MAPK signaling pathway for regulating host defense (Figure 2) and counterparts of TAK1 (41), MKK3/4 (42), p38 (42), and JNK (43) have been cloned and demonstrated to play a role in innate immune responses. It will be interesting to determine whether their activation depends on signaling through Toll and Tube.
Surprisingly, TLR2 was recently found to signal apoptosis in response to stimulation by BLPs and LPS (22, 44), a situation reminiscent of TNF-receptor signaling, in which cell activation and death can be executed through a single receptor (45). However, the molecular identity of signaling components involved in TLR2-induced apoptosis is currently unknown. Apoptosis through TLRs may play a role in the resolution of inflammation by limiting the number of activated cells that produce a large amount of proinflammatory mediators (22). It will be interesting to determine when and how life and death decisions occur during an infection.
NF-κB and Rel proteins in innate and adaptive immunity
Given the diverse nature of signals that impinge on NF-κB, and the variety of genes that it regulates, it is not difficult to appreciate the importance of NF-κB in different aspects of normal physiology. Under normal conditions, NF-κB is rapidly activated upon microbial invasion, and this activation usually correlates with the resistance of the host to infection. However, persistent activation of NF-κB may lead to the production of excessive amounts of proinflammatory mediators, resulting in tissue damage, organ failure, and even death of the host, as in bacterial infection–induced septic shock. It is interesting to note that in order to survive in the host, certain pathogens, such as Bordetella (46), Yersinia (47), and African swine fever virus (48), have evolved mechanisms to counteract or escape the host immune system by inhibiting NF-κB activation. On the other hand, some viruses, including HIV-1, CMV, and SV-40, take advantage of NF-κB, a host factor that is activated at sites of infection, for transcription of their own genes and enhanced infectivity (6, 7). Therefore, modulating NF-κB activity appears to be a very attractive strategy for treating various infectious and inflammatory diseases (see Hiscott, this series, ref. 49).
Recent genetic evidence with knockout and transgenic mice has reinforced the importance of individual NF-κB/Rel proteins in innate and adaptive immune responses, as well as in development and cell survival (reviewed in ref. 6). Thus far, all five NF-κB/Rel proteins have been deleted and in some cases, double knockouts have also been generated (6). Collectively, these data indicate that individual Rel proteins play important roles in innate and adaptive immune responses, although their contributions vary among the different members.
Summary and future prospects
The discovery of TLRs has opened up a wide area for further study. It is now quite clear that they are the long-sought-after CD14-associated signaling transducers for LPS and other microbial products. However, a key question that remains to be answered concerns the roles of the various TLRs in innate immunity. Do individual members recognize distinct microbial products (the PAMPs), thus linking individual TLRs to particular pathogens and thereby altering the perception of innate immunity as being a strictly nonspecific recognition system? Recent findings with TLR2 and TLR4 suggest that such specificity between individual TLRs exists, but more members of this family will need to be studied in order to obtain a more complete picture of how specific pattern recognition occurs in innate immune responses. An additional twist to the functioning of TLRs is suggested by the nature of Toll-related proteins that are found in plants, such as the N protein. Although these proteins resemble the TLRs, they are exclusively intracellular and hence most likely used to recognize intracellular pathogens. Similar intracellular receptors probably exist in mammalian cells and their identification and characterization will be an important area of future investigation.
Another important question posed by the discovery of the TLRs and the demonstration of their role in innate immunity concerns the signal transduction pathways activated by these receptors. The phenotype of different knockout animals suggests that TLRs do not signal exclusively through MyD88/IRAK-dependent pathways. Therefore the characterization of alternate pathways and signaling intermediates leading from TLRs to NF-κB or AP-1 is likely to be an active area of investigation. Although evolutionary considerations strongly implicate NF-κB and AP-1 as critical effectors of TLR-initiated innate immune responses, it remains unclear whether the activation of these transcription factors is sufficient to mount the complete innate immune response. It is also unclear whether all TLRs can activate NF-κB and AP-1 or whether some TLRs interact with alternative signaling pathways. Finally the recent demonstration that exposure to BLPs can lead to apoptosis makes the TLR similar to other cytokine receptors that can signal either activation or death. The molecular events that determine this choice remain unclear, however the elucidation of the details of this switch will be of great interest, since the killing of activated cells at sites of infection or inflammation might be a critical step in the resolution of innate immune responses.
The discoveries over the past few years have resulted in a significant expansion of our knowledge of mechanisms of innate immune recognition and the impact of innate immunity on adaptive immune responses. Innate immunity is gaining renewed interest from immunologists and microbiologists, and a better understanding of this signaling pathway will certainly lead to more effective therapeutic strategies for treating various infections, as well as acute and chronic inflammatory diseases, such as septic shock, rheumatoid arthritis, and inflammatory bowel disease.
Acknowledgments
Work in the authors’ laboratory is supported by the Howard Hughes Medical Institute and the NIH. We would like to thank Ruslan Medzhitov and Michael May for thoughtful suggestions and careful reading of the manuscript. G. Zhang would like to thank Frank Blecha for continuous encouragement and support. We apologize for failing to refer to many primary sources due to space constraints.
References
- 1.Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–53. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Janeway CA., Jr Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 4.Janeway CA, Jr, Medzhitov R. Lipoproteins take their toll on the host. Curr Biol. 1999;9:R879–R882. doi: 10.1016/s0960-9822(00)80073-1. [DOI] [PubMed] [Google Scholar]
- 5.Anderson KV. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 7.May MJ, Ghosh S. Signal transduction through NF-κB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 8.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 9.Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 10.O’Neill LA, Greene C. Signal transduction pathways activated by the IL-1 receptor family: ancient signaling machinery in mammals, insects, and plants. J Leukoc Biol. 1998;63:650–657. [PubMed] [Google Scholar]
- 11.Diamond G, Kaiser V, Rhodes J, Russell JP, Bevins CL. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect Immun. 2000;68:113–119. doi: 10.1128/iai.68.1.113-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkett M, Gilmore TD. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene. 1999;18:6910–6924. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- 13.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 14.Muzio M, Polentarutti N, Bosisio D, Prahladan MK, Mantovani A. Toll-like receptors: a growing family of immune receptors that are differentially expressed and regulated by different leukocytes. J Leukoc Biol. 2000;67:450–456. doi: 10.1002/jlb.67.4.450. [DOI] [PubMed] [Google Scholar]
- 15.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 16.Fenton MJ, Golenbock DT. LPS-binding proteins and receptors. J Leukoc Biol. 1998;64:25–32. doi: 10.1002/jlb.64.1.25. [DOI] [PubMed] [Google Scholar]
- 17.Beutler B. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12:20–26. doi: 10.1016/s0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- 18.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 19.Yang RB, et al. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 20.Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang RB, Mark MR, Gurney AL, Godowski PJ. Signaling events induced by lipopolysaccharide-activated toll-like receptor 2. J Immunol. 1999;163:639–643. [PubMed] [Google Scholar]
- 22.Aliprantis AO, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 23.Brightbill HD, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 24.Hoshino K, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 25.Arbour NC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 26.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 28.Lien E, et al. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poltorak A, Ricciardi-Castagnoli P, Citterio S, Beutler B. Physical contact between lipopolysaccharide and toll-like receptor 4 revealed by genetic complementation. Proc Natl Acad Sci USA. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi K, Burkart V, Flohe S, Kolb H. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol. 2000;164:558–561. doi: 10.4049/jimmunol.164.2.558. [DOI] [PubMed] [Google Scholar]
- 31.Asea A, et al. HSP70 stimulates cytokine production through a CD14-dependent pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 32.Kopp E, et al. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninomiya-Tsuji J, et al. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 34.Takaesu G, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 35.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Su YC, Becker E, Treisman J, Skolnik EY. A Drosophila TNF-receptor-associated factor (TRAF) binds the ste20 kinase Misshapen and activates Jun kinase. Curr Biol. 1999;9:101–104. doi: 10.1016/s0960-9822(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 37.Zapata JM, et al. The Drosophila tumor necrosis factor receptor-associated factor-1 (DTRAF1) interacts with Pelle and regulates NFkappaB activity. J Biol Chem. 2000;275:12102–12107. doi: 10.1074/jbc.275.16.12102. [DOI] [PubMed] [Google Scholar]
- 38.Kim YS, et al. Lipopolysaccharide-activated kinase, an essential component for the induction of the antimicrobial peptide genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:2071–2079. doi: 10.1074/jbc.275.3.2071. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Young DW, Gusovsky F, Chow JC. Cellular events mediated by lipopolysaccharide-stimulated toll-like receptor 4. MD-2 is required for activation of mitogen-activated protein kinases and Elk-1. J Biol Chem. 2000;275:20861–20866. doi: 10.1074/jbc.M002896200. [DOI] [PubMed] [Google Scholar]
- 40.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 41.Takatsu Y, et al. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol Cell Biol. 2000;20:3015–3026. doi: 10.1128/mcb.20.9.3015-3026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han ZS, et al. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluss HK, Han Z, Barrett T, Davis RJ, Ip YT. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 44.Aliprantis AO, Yang RB, Weiss DS, Godowski P, Zychlinsky A. The apoptotic signaling pathway activated by Toll-like receptor-2. EMBO J. 2000;19:3325–3336. doi: 10.1093/emboj/19.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallach D, et al. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 46.Yuk MH, Harvill ET, Cotter PA, Miller JF. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by the Bordetella type III secretion system. Mol Microbiol. 2000;35:991–1004. doi: 10.1046/j.1365-2958.2000.01785.x. [DOI] [PubMed] [Google Scholar]
- 47.Ruckdeschel K, et al. Yersinia enterocolitica impairs activation of transcription factor NF-κB: involvement in the induction of programmed cell death and in the suppression of the macrophage tumor necrosis factor alpha production. J Exp Med. 1998;187:1069–1079. doi: 10.1084/jem.187.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powell PP, Dixon LK, Parkhouse RM. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J Virol. 1996;70:8527–8533. doi: 10.1128/jvi.70.12.8527-8533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiscott, J., Kwon, H., and Génin, P. 2001. Hostile takeovers: viral appropriation of the NF-κB pathway. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]